| 1 |
|
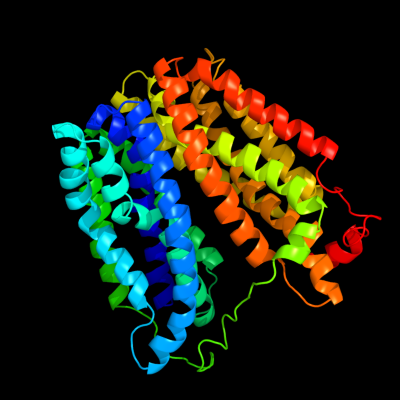
PDB 1pv7 chain A
Region: 9 - 457
Aligned: 413
Modelled: 434
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 2 |
|
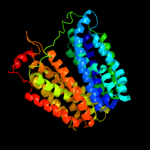
PDB 1pw4 chain A
Region: 2 - 448
Aligned: 414
Modelled: 432
Confidence: 100.0%
Identity: 14%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
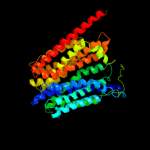
PDB 3o7p chain A
Region: 19 - 437
Aligned: 382
Modelled: 404
Confidence: 99.9%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 4 |
|
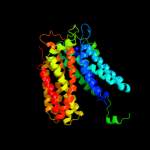
PDB 2gfp chain A
Region: 16 - 436
Aligned: 372
Modelled: 394
Confidence: 99.9%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 21 - 425
Aligned: 387
Modelled: 405
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 375 - 453
Aligned: 79
Modelled: 79
Confidence: 76.6%
Identity: 13%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 3hd6 chain A
Region: 220 - 460
Aligned: 220
Modelled: 241
Confidence: 54.3%
Identity: 10%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 8 |
|
PDB 3d9s chain B
Region: 413 - 454
Aligned: 42
Modelled: 42
Confidence: 20.9%
Identity: 14%
PDB header:membrane protein
Chain: B: PDB Molecule:aquaporin-5;
PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
Phyre2
| 9 |
|
PDB 3b9y chain A
Region: 223 - 463
Aligned: 212
Modelled: 224
Confidence: 16.3%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 10 |
|
PDB 2oar chain A domain 1
Region: 413 - 467
Aligned: 45
Modelled: 55
Confidence: 15.0%
Identity: 18%
Fold: Gated mechanosensitive channel
Superfamily: Gated mechanosensitive channel
Family: Gated mechanosensitive channel
Phyre2
| 11 |
|
PDB 2w8a chain C
Region: 313 - 461
Aligned: 130
Modelled: 149
Confidence: 10.5%
Identity: 9%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 12 |
|
PDB 1by0 chain A
Region: 438 - 463
Aligned: 26
Modelled: 26
Confidence: 9.3%
Identity: 19%
PDB header:rna binding protein
Chain: A: PDB Molecule:protein (hepatitis delta antigen);
PDBTitle: n-terminal leucine-repeat region of hepatitis delta antigen
Phyre2
| 13 |
|
PDB 1ymg chain A
Region: 413 - 449
Aligned: 37
Modelled: 37
Confidence: 7.0%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
Phyre2
| 14 |
|
PDB 1ymg chain A domain 1
Region: 413 - 449
Aligned: 37
Modelled: 37
Confidence: 7.0%
Identity: 14%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 15 |
|
PDB 1j4n chain A
Region: 413 - 447
Aligned: 35
Modelled: 35
Confidence: 6.5%
Identity: 14%
Fold: Aquaporin-like
Superfamily: Aquaporin-like
Family: Aquaporin-like
Phyre2
| 16 |
|
PDB 2oar chain A
Region: 413 - 467
Aligned: 45
Modelled: 55
Confidence: 6.3%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2
| 17 |
|
PDB 1xao chain A
Region: 434 - 467
Aligned: 34
Modelled: 34
Confidence: 5.7%
Identity: 12%
PDB header:chaperone
Chain: A: PDB Molecule:mitochondrial protein import protein mas5;
PDBTitle: hsp40-ydj1 dimerization domain
Phyre2