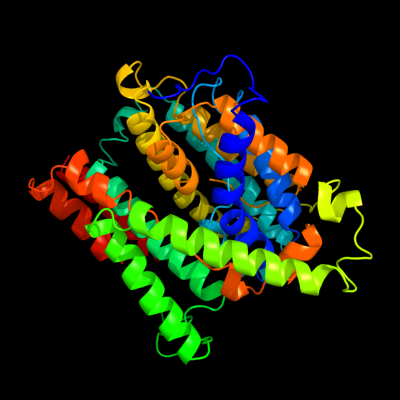
1 c3qe7A_
100.0
13
PDB header: transport proteinChain: A: PDB Molecule: uracil permease;PDBTitle: crystal structure of uracil transporter--uraa
2 c3lpzA_
25.2
19
PDB header: protein transportChain: A: PDB Molecule: get4 (yor164c homolog);PDBTitle: crystal structure of c. therm. get4
3 d1v54l_
19.1
24
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)Family: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)4 c3iysA_
16.5
47
PDB header: virusChain: A: PDB Molecule: major capsid protein vp1;PDBTitle: homology model of avian polyomavirus asymmetric unit
5 c1w3gA_
13.3
29
PDB header: toxin/lectinChain: A: PDB Molecule: hemolytic lectin from laetiporus sulphureus;PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
6 c2x7lP_
11.2
27
PDB header: immune systemChain: P: PDB Molecule: hiv rev;PDBTitle: implications of the hiv-1 rev dimer structure at 3.2a2 resolution for multimeric binding to the rev response3 element
7 c2y69Y_
10.7
25
PDB header: electron transportChain: Y: PDB Molecule: cytochrome c oxidase subunit 7c;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
8 d1sida_
10.3
50
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group I dsDNA virusesFamily: Papovaviridae-like VP9 d1sva1_
9.9
53
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group I dsDNA virusesFamily: Papovaviridae-like VP10 c3lphD_
9.0
31
PDB header: viral proteinChain: D: PDB Molecule: protein rev;PDBTitle: crystal structure of the hiv-1 rev dimer
11 c1hynQ_
8.6
36
PDB header: membrane proteinChain: Q: PDB Molecule: band 3 anion transport protein;PDBTitle: crystal structure of the cytoplasmic domain of human2 erythrocyte band-3 protein
12 d3er7a1
8.4
28
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Exig0174-like13 d1hynp_
8.4
36
Fold: Phoshotransferase/anion transport proteinSuperfamily: Phoshotransferase/anion transport proteinFamily: Anion transport protein, cytoplasmic domain14 c2elpA_
7.5
32
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
15 c3hfxA_
7.5
9
PDB header: transport proteinChain: A: PDB Molecule: l-carnitine/gamma-butyrobetaine antiporter;PDBTitle: crystal structure of carnitine transporter
16 c2kkuA_
7.4
42
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of protein af2351 from archaeoglobus2 fulgidus. northeast structural genomics consortium target3 att9/ontario center for structural proteomics target af2351
17 d1pv0a_
7.4
21
Fold: Long alpha-hairpinSuperfamily: Sporulation inhibitor SdaFamily: Sporulation inhibitor Sda18 c3dinD_
7.2
10
PDB header: membrane protein, protein transportChain: D: PDB Molecule: preprotein translocase subunit sece;PDBTitle: crystal structure of the protein-translocation complex formed by the2 secy channel and the seca atpase
19 c3kihC_
7.1
23
PDB header: sugar binding proteinChain: C: PDB Molecule: 5-bladed -propeller lectin;PDBTitle: the crystal structures of two fragments truncated from 5-bladed -2 propeller lectin, tachylectin-2 (lib2-d2-15)
20 d1txna_
6.7
43
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase21 d2gr7a1
not modelled
6.5
18
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: YadA C-terminal domain-like22 c2gr7C_
not modelled
6.5
18
PDB header: membrane proteinChain: C: PDB Molecule: adhesin;PDBTitle: hia 992-1098
23 c2ziiA_
not modelled
6.5
21
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associated protein 74;PDBTitle: crystal structure of yeast vps74-n-term truncation variant
24 c2zihC_
not modelled
6.4
21
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 74;PDBTitle: crystal structure of yeast vps74
25 c2aexA_
not modelled
6.3
43
PDB header: oxidoreductaseChain: A: PDB Molecule: coproporphyrinogen iii oxidase, mitochondrial;PDBTitle: the 1.58a crystal structure of human coproporphyrinogen oxidase2 reveals the structural basis of hereditary coproporphyria
26 c3ns4A_
not modelled
6.2
14
PDB header: protein bindingChain: A: PDB Molecule: vacuolar protein sorting-associated protein 53;PDBTitle: structure of a c-terminal fragment of its vps53 subunit suggests2 similarity of garp to a family of tethering complexes
27 c1u7bB_
not modelled
6.1
35
PDB header: replicationChain: B: PDB Molecule: srqgstqgrlddffkvtgsl peptide of flapPDBTitle: crystal structure of hpcna bound to residues 331-350 of the2 flap endonuclease-1 (fen1)
28 c3hymA_
not modelled
6.1
17
PDB header: cell cycle, ligaseChain: A: PDB Molecule: anaphase-promoting complex subunit cdc26;PDBTitle: insights into anaphase promoting complex tpr subdomain2 assembly from a cdc26-apc6 structure
29 c3hymE_
not modelled
6.1
17
PDB header: cell cycle, ligaseChain: E: PDB Molecule: anaphase-promoting complex subunit cdc26;PDBTitle: insights into anaphase promoting complex tpr subdomain2 assembly from a cdc26-apc6 structure
30 c3hymI_
not modelled
6.1
17
PDB header: cell cycle, ligaseChain: I: PDB Molecule: anaphase-promoting complex subunit cdc26;PDBTitle: insights into anaphase promoting complex tpr subdomain2 assembly from a cdc26-apc6 structure
31 d1tkla_
not modelled
6.0
43
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase32 c3omdB_
not modelled
6.0
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of unknown function protein from leptospirillum2 rubarum
33 c2ogfD_
not modelled
5.9
36
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: hypothetical protein mj0408;PDBTitle: crystal structure of protein mj0408 from methanococcus jannaschii,2 pfam duf372
34 d1vjua_
not modelled
5.9
35
Fold: Coproporphyrinogen III oxidaseSuperfamily: Coproporphyrinogen III oxidaseFamily: Coproporphyrinogen III oxidase35 d1vqoi1
not modelled
5.8
32
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein L11, C-terminal domainFamily: Ribosomal protein L11, C-terminal domain36 c1vq8I_
not modelled
5.8
32
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: the structure of ccda-phe-cap-bio and the antibiotic sparsomycin bound2 to the large ribosomal subunit of haloarcula marismortui
37 c1bmxA_
not modelled
5.6
37
PDB header: viral proteinChain: A: PDB Molecule: human immunodeficiency virus type 1 capsid;PDBTitle: hiv-1 capsid protein major homology region peptide analog,2 nmr, 8 structures
38 c2ht2B_
not modelled
5.5
13
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
39 c1zv8I_
not modelled
5.3
7
PDB header: viral proteinChain: I: PDB Molecule: e2 glycoprotein;PDBTitle: a structure-based mechanism of sars virus membrane fusion
40 d1avwb_
not modelled
5.3
21
Fold: beta-TrefoilSuperfamily: STI-likeFamily: Kunitz (STI) inhibitors41 c1btqA_
not modelled
5.2
18
PDB header: anion transportChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
42 c1btrA_
not modelled
5.2
18
PDB header: anion transportChain: A: PDB Molecule: band 3 anion transport protein;PDBTitle: the solution structures of the first and second2 transmembrane-spanning segments of band 3
43 d1q90m_
not modelled
5.2
32
Fold: Single transmembrane helixSuperfamily: PetM subunit of the cytochrome b6f complexFamily: PetM subunit of the cytochrome b6f complex