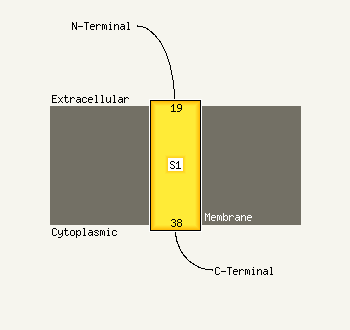
1 c3fqmA_
39.8
27
PDB header: metal binding proteinChain: A: PDB Molecule: non-structural protein 5a;PDBTitle: crystal structure of a novel dimeric form of hcv ns5a domain i protein
2 d1e2ta_
33.5
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase3 c2pfrB_
25.5
14
PDB header: transferaseChain: B: PDB Molecule: arylamine n-acetyltransferase 2;PDBTitle: human n-acetyltransferase 2
4 c3lnbA_
24.4
9
PDB header: transferaseChain: A: PDB Molecule: n-acetyltransferase family protein;PDBTitle: crystal structure analysis of arylamine n-acetyltransferase c from2 bacillus anthracis
5 c2qtxL_
22.3
16
PDB header: rna binding proteinChain: L: PDB Molecule: uncharacterized protein mj1435;PDBTitle: crystal structure of an hfq-like protein from methanococcus2 jannaschii
6 c1j3wB_
20.4
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: giding protein-mglb;PDBTitle: structure of gliding protein-mglb from thermus thermophilus hb8
7 c3hh1D_
19.9
12
PDB header: transferaseChain: D: PDB Molecule: tetrapyrrole methylase family protein;PDBTitle: the structure of a tetrapyrrole methylase family protein domain from2 chlorobium tepidum tls
8 c2kncA_
18.4
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
9 c2l2tA_
16.6
32
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-4;PDBTitle: solution nmr structure of the erbb4 dimeric membrane domain
10 d1j3wa_
16.3
22
Fold: Profilin-likeSuperfamily: Roadblock/LC7 domainFamily: Roadblock/LC7 domain11 d1pjqa2
15.8
24
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase12 c3e19D_
14.1
19
PDB header: transcription regulator, metal binding pChain: D: PDB Molecule: feoa;PDBTitle: crystal structure of iron uptake regulatory protein (feoa) solved by2 sulfur sad in a monoclinic space group
13 c2e0kA_
13.9
12
PDB header: transferaseChain: A: PDB Molecule: precorrin-2 c20-methyltransferase;PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
14 c1xe1A_
13.1
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pf0907;PDBTitle: hypothetical protein from pyrococcus furiosus pfu-880080-001
15 d1xe1a_
13.1
15
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Elongation factors16 d1ve2a1
12.8
18
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase17 c1v55B_
12.8
0
PDB header: oxidoreductaseChain: B: PDB Molecule: cytochrome c oxidase polypeptide ii;PDBTitle: bovine heart cytochrome c oxidase at the fully reduced state
18 c3gztF_
12.6
50
PDB header: virusChain: F: PDB Molecule: outer capsid glycoprotein vp7;PDBTitle: vp7 recoated rotavirus dlp
19 c3kwpA_
12.5
6
PDB header: transferaseChain: A: PDB Molecule: predicted methyltransferase;PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
20 d1fftb2
12.5
19
Fold: Transmembrane helix hairpinSuperfamily: Cytochrome c oxidase subunit II-like, transmembrane regionFamily: Cytochrome c oxidase subunit II-like, transmembrane region21 c1afoB_
not modelled
12.4
25
PDB header: integral membrane proteinChain: B: PDB Molecule: glycophorin a;PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
22 c3nutC_
not modelled
12.2
24
PDB header: transferaseChain: C: PDB Molecule: precorrin-3 methylase;PDBTitle: crystal structure of the methyltransferase cobj
23 d1va0a1
not modelled
12.2
24
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase24 c2zvbA_
not modelled
12.0
18
PDB header: transferaseChain: A: PDB Molecule: precorrin-3 c17-methyltransferase;PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
25 c2qbuA_
not modelled
11.7
29
PDB header: transferaseChain: A: PDB Molecule: precorrin-2 methyltransferase;PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
26 c2jwaA_
not modelled
11.6
11
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
27 c2ks1B_
not modelled
11.0
23
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
28 d1q18a1
not modelled
10.9
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glucokinase29 c1pjtB_
not modelled
10.8
24
PDB header: transferase/oxidoreductase/lyaseChain: B: PDB Molecule: siroheme synthase;PDBTitle: the structure of the ser128ala point-mutant variant of cysg,2 the multifunctional3 methyltransferase/dehydrogenase/ferrochelatase for4 siroheme synthesis
30 c2yboA_
not modelled
10.5
24
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
31 d1s4da_
not modelled
9.7
24
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase32 c1cbfA_
not modelled
9.5
18
PDB header: methyltransferaseChain: A: PDB Molecule: cobalt-precorrin-4 transmethylase;PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
33 d1cbfa_
not modelled
9.5
18
Fold: Tetrapyrrole methylaseSuperfamily: Tetrapyrrole methylaseFamily: Tetrapyrrole methylase34 d1ppjd2
not modelled
9.2
17
Fold: Single transmembrane helixSuperfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchorFamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor35 d1vdda_
not modelled
9.1
23
Fold: Recombination protein RecRSuperfamily: Recombination protein RecRFamily: Recombination protein RecR36 c3q1jA_
not modelled
8.5
8
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20;PDBTitle: crystal structure of tudor domain 1 of human phd finger protein 20
37 c3kdpG_
not modelled
8.5
33
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
38 c3kdpH_
not modelled
8.5
33
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
39 c3sftA_
not modelled
8.4
21
PDB header: hydrolaseChain: A: PDB Molecule: chemotaxis response regulator protein-glutamatePDBTitle: crystal structure of thermotoga maritima cheb methylesterase catalytic2 domain
40 c2i2rK_
not modelled
8.3
18
PDB header: transport proteinChain: K: PDB Molecule: potassium voltage-gated channel subfamily d member 3;PDBTitle: crystal structure of the kchip1/kv4.3 t1 complex
41 d3cx5d2
not modelled
8.1
13
Fold: Single transmembrane helixSuperfamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchorFamily: Cytochrome c1 subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase), transmembrane anchor42 d2ftwa1
not modelled
7.6
25
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Hydantoinase (dihydropyrimidinase)43 c2fynH_
not modelled
7.5
24
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
44 c2r6fA_
not modelled
7.4
19
PDB header: hydrolaseChain: A: PDB Molecule: excinuclease abc subunit a;PDBTitle: crystal structure of bacillus stearothermophilus uvra
45 c2l34A_
not modelled
7.4
25
PDB header: protein bindingChain: A: PDB Molecule: tyro protein tyrosine kinase-binding protein;PDBTitle: structure of the dap12 transmembrane homodimer
46 c2l34B_
not modelled
7.2
26
PDB header: protein bindingChain: B: PDB Molecule: tyro protein tyrosine kinase-binding protein;PDBTitle: structure of the dap12 transmembrane homodimer
47 d1w4ta1
not modelled
6.6
13
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase48 d2bsza1
not modelled
6.4
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Arylamine N-acetyltransferase49 c2l35A_
not modelled
6.4
29
PDB header: protein bindingChain: A: PDB Molecule: dap12-nkg2c_tm;PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
50 c2kpeA_
not modelled
6.4
29
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
51 c2kpeB_
not modelled
6.4
29
PDB header: membrane proteinChain: B: PDB Molecule: glycophorin-a;PDBTitle: refined structure of glycophorin a transmembrane segment dimer in dpc2 micelles
52 c3cwbQ_
not modelled
6.2
10
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
53 c2npnA_
not modelled
6.2
25
PDB header: transferaseChain: A: PDB Molecule: putative cobalamin synthesis related protein;PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
54 c2rmzA_
not modelled
5.7
32
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
55 c2k21A_
not modelled
5.7
24
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
56 c2yiuE_
not modelled
5.6
24
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
57 c3nd1B_
not modelled
5.5
13
PDB header: transferaseChain: B: PDB Molecule: precorrin-6a synthase/cobf protein;PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
58 d2j01v1
not modelled
5.4
6
Fold: L21p-likeSuperfamily: L21p-likeFamily: Ribosomal protein L21p59 c2rddB_
not modelled
5.4
13
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
60 c1vddC_
not modelled
5.1
23
PDB header: recombinationChain: C: PDB Molecule: recombination protein recr;PDBTitle: crystal structure of recombinational repair protein recr
61 c1p84D_
not modelled
5.1
24
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex