| 1 |
|
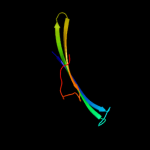
PDB 1osm chain A
Region: 22 - 92
Aligned: 71
Modelled: 71
Confidence: 99.8%
Identity: 63%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Porin
Phyre2
| 2 |
|
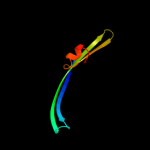
PDB 1pho chain A
Region: 22 - 92
Aligned: 71
Modelled: 71
Confidence: 99.8%
Identity: 58%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Porin
Phyre2
| 3 |
|
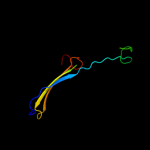
PDB 3nsg chain A
Region: 23 - 93
Aligned: 71
Modelled: 71
Confidence: 99.8%
Identity: 51%
PDB header:membrane protein
Chain: A: PDB Molecule:outer membrane protein f;
PDBTitle: crystal structure of ompf, an outer membrane protein from salmonella2 typhi
Phyre2
| 4 |
|
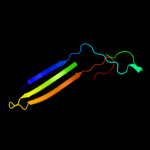
PDB 2zfg chain A domain 1
Region: 22 - 94
Aligned: 73
Modelled: 73
Confidence: 99.8%
Identity: 58%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Porin
Phyre2
| 5 |
|
PDB 2fgq chain X domain 1
Region: 31 - 95
Aligned: 65
Modelled: 65
Confidence: 99.6%
Identity: 20%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Porin
Phyre2
| 6 |
|
PDB 3a2r chain X
Region: 32 - 94
Aligned: 63
Modelled: 63
Confidence: 99.6%
Identity: 17%
PDB header:membrane protein
Chain: X: PDB Molecule:outer membrane protein ii;
PDBTitle: crystal structure of outer membrane protein porb from neisseria2 meningitidis
Phyre2
| 7 |
|
PDB 3prn chain A
Region: 32 - 94
Aligned: 63
Modelled: 63
Confidence: 99.2%
Identity: 13%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Porin
Phyre2
| 8 |
|
PDB 2por chain A
Region: 32 - 91
Aligned: 58
Modelled: 60
Confidence: 99.2%
Identity: 10%
Fold: Transmembrane beta-barrels
Superfamily: Porins
Family: Porin
Phyre2
| 9 |
|
PDB 2o4v chain A
Region: 26 - 88
Aligned: 61
Modelled: 63
Confidence: 90.1%
Identity: 23%
PDB header:membrane protein
Chain: A: PDB Molecule:porin p;
PDBTitle: an arginine ladder in oprp mediates phosphate specific transfer across2 the outer membrane
Phyre2
| 10 |
|
PDB 2iwv chain D
Region: 30 - 90
Aligned: 61
Modelled: 61
Confidence: 71.6%
Identity: 11%
PDB header:ion channel
Chain: D: PDB Molecule:outer membrane protein g;
PDBTitle: structure of the monomeric outer membrane porin ompg in the2 open and closed conformation
Phyre2
| 11 |
|
PDB 2jna chain A domain 1
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 62.9%
Identity: 25%
Fold: Dodecin subunit-like
Superfamily: YdgH-like
Family: YdgH-like
Phyre2
| 12 |
|
PDB 1uyn chain X
Region: 29 - 84
Aligned: 56
Modelled: 56
Confidence: 11.3%
Identity: 14%
Fold: Transmembrane beta-barrels
Superfamily: Autotransporter
Family: Autotransporter
Phyre2
| 13 |
|
PDB 2wjq chain A
Region: 58 - 95
Aligned: 38
Modelled: 38
Confidence: 10.1%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:probable n-acetylneuraminic acid outer membrane channel
PDBTitle: nanc porin structure in hexagonal crystal form.
Phyre2
| 14 |
|
PDB 2vda chain B
Region: 2 - 21
Aligned: 20
Modelled: 20
Confidence: 9.9%
Identity: 25%
PDB header:protein transport
Chain: B: PDB Molecule:maltoporin;
PDBTitle: solution structure of the seca-signal peptide complex
Phyre2
| 15 |
|
PDB 2jwy chain A
Region: 36 - 80
Aligned: 40
Modelled: 45
Confidence: 6.4%
Identity: 23%
PDB header:lipoprotein
Chain: A: PDB Molecule:uncharacterized lipoprotein yaji;
PDBTitle: solution nmr structure of uncharacterized lipoprotein yaji from2 escherichia coli. northeast structural genomics target er540
Phyre2