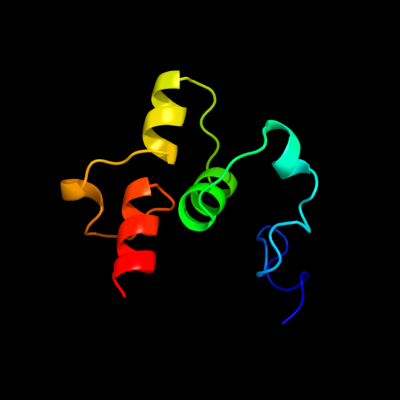
1 c3knyA_
53.1
21
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein bt_3535;PDBTitle: crystal structure of a two domain protein with unknown function2 (bt_3535) from bacteroides thetaiotaomicron vpi-5482 at 2.60 a3 resolution
2 c1pbyA_
48.8
21
PDB header: oxidoreductaseChain: A: PDB Molecule: quinohemoprotein amine dehydrogenase 60 kdaPDBTitle: structure of the phenylhydrazine adduct of the2 quinohemoprotein amine dehydrogenase from paracoccus3 denitrificans at 1.7 a resolution
3 d1qmga1
45.0
20
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: Acetohydroxy acid isomeroreductase (ketol-acid reductoisomerase, KARI)4 c2v79B_
29.4
12
PDB header: dna-binding proteinChain: B: PDB Molecule: dna replication protein dnad;PDBTitle: crystal structure of the n-terminal domain of dnad from2 bacillus subtilis
5 d1khba1
27.5
31
Fold: PEP carboxykinase-likeSuperfamily: PEP carboxykinase-likeFamily: PEP carboxykinase C-terminal domain6 c2vn2B_
24.5
14
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
7 c2qeyA_
23.8
31
PDB header: lyaseChain: A: PDB Molecule: phosphoenolpyruvate carboxykinase, cytosolic [gtp];PDBTitle: rat cytosolic pepck in complex with gtp
8 c2f5uA_
20.6
30
PDB header: viral proteinChain: A: PDB Molecule: virion protein ul25;PDBTitle: structural characterization of the ul25 dna packaging2 protein from herpes simplex virus type 1
9 c2jrdA_
20.6
31
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin;PDBTitle: influenza hemagglutinin fusion domain mutant f9a
10 d1bdta_
19.1
27
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors11 c2zciD_
18.5
38
PDB header: signaling protein, lyaseChain: D: PDB Molecule: phosphoenolpyruvate carboxykinase [gtp];PDBTitle: structure of a gtp-dependent bacterial pep-carboxykinase2 from corynebacterium glutamicum
12 d1t07a_
17.2
20
Fold: YggX-likeSuperfamily: YggX-likeFamily: YggX-like13 d1xs8a_
16.1
17
Fold: YggX-likeSuperfamily: YggX-likeFamily: YggX-like14 c1jmxA_
15.9
24
PDB header: oxidoreductaseChain: A: PDB Molecule: amine dehydrogenase;PDBTitle: crystal structure of a quinohemoprotein amine dehydrogenase2 from pseudomonas putida
15 d1seda_
15.9
17
Fold: Hypothetical protein YhaISuperfamily: Hypothetical protein YhaIFamily: Hypothetical protein YhaI16 c2fahB_
13.2
25
PDB header: lyaseChain: B: PDB Molecule: phosphoenolpyruvate carboxykinase;PDBTitle: the structure of mitochondrial pepck, complex with mn and gdp
17 c3bdrA_
12.6
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ycf58 protein;PDBTitle: crystal structure of fatty acid-binding protein-like ycf582 from thermosynecoccus elongatus. northeast structural3 genomics consortium target ter13.
18 d1pbya1
12.5
29
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 219 c1tw3A_
12.0
13
PDB header: transferaseChain: A: PDB Molecule: carminomycin 4-o-methyltransferase;PDBTitle: crystal structure of carminomycin-4-o-methyltransferase2 (dnrk) in complex with s-adenosyl-l-homocystein (sah) and3 4-methoxy-e-rhodomycin t (m-et)
20 d1gjja2
11.3
17
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain21 d6paxa1
not modelled
11.2
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain22 c1iboA_
not modelled
10.5
31
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin ha2 chain peptide;PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 7.4
23 c1ibnA_
not modelled
10.5
31
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin ha2 chain peptide;PDBTitle: nmr structure of hemagglutinin fusion peptide in dpc2 micelles at ph 5
24 d1z5ga1
not modelled
10.2
12
Fold: HAD-likeSuperfamily: HAD-likeFamily: Class B acid phosphatase, AphA25 d1zbsa2
not modelled
9.5
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like26 c3chxG_
not modelled
9.3
23
PDB header: membrane proteinChain: G: PDB Molecule: pmoc;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
27 c2kxaA_
not modelled
9.1
23
PDB header: viral protein, immune systemChain: A: PDB Molecule: haemagglutinin ha2 chain peptide;PDBTitle: the hemagglutinin fusion peptide (h1 subtype) at ph 7.4
28 c1gjjA_
not modelled
9.1
33
PDB header: membrane proteinChain: A: PDB Molecule: lap2;PDBTitle: n-terminal constant region of the nuclear envelope protein2 lap2
29 d1b28a_
not modelled
8.8
29
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors30 d1h9fa_
not modelled
8.2
30
Fold: LEM/SAP HeH motifSuperfamily: LEM domainFamily: LEM domain31 d1k78a1
not modelled
7.8
25
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain32 d1iuqa_
not modelled
7.7
17
Fold: Glycerol-3-phosphate (1)-acyltransferaseSuperfamily: Glycerol-3-phosphate (1)-acyltransferaseFamily: Glycerol-3-phosphate (1)-acyltransferase33 c1yewC_
not modelled
7.0
15
PDB header: oxidoreductase, membrane proteinChain: C: PDB Molecule: particulate methane monooxygenase subunit c2;PDBTitle: crystal structure of particulate methane monooxygenase
34 c2pjwH_
not modelled
6.8
16
PDB header: endocytosis/exocytosisChain: H: PDB Molecule: uncharacterized protein yhl002w;PDBTitle: the vps27/hse1 complex is a gat domain-based scaffold for2 ubiquitin-dependent sorting
35 c2k1oA_
not modelled
6.7
41
PDB header: gene regulationChain: A: PDB Molecule: putative;PDBTitle: nmr structure of helicobacter pylori jhp0511 (hp0564).
36 c2pfcA_
not modelled
6.6
22
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein rv0098/mt0107;PDBTitle: structure of mycobacterium tuberculosis rv0098
37 c1xopA_
not modelled
6.4
31
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin;PDBTitle: nmr structure of g1v mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
38 c1xooA_
not modelled
6.3
31
PDB header: viral proteinChain: A: PDB Molecule: hemagglutinin;PDBTitle: nmr structure of g1s mutant of influenza hemagglutinin2 fusion peptide in dpc micelles at ph 5
39 c3bhwA_
not modelled
6.1
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein from magnetospirillum2 magneticum
40 d1fi6a_
not modelled
5.9
24
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)41 d1c07a_
not modelled
5.7
19
Fold: EF Hand-likeSuperfamily: EF-handFamily: Eps15 homology domain (EH domain)42 c2bn5A_
not modelled
5.6
25
PDB header: nuclear proteinChain: A: PDB Molecule: psi;PDBTitle: p-element somatic inhibitor protein complex with u1-70k2 proline-rich peptide
43 c2ia2D_
not modelled
5.5
13
PDB header: transcriptionChain: D: PDB Molecule: putative transcriptional regulator;PDBTitle: the crystal structure of a putative transcriptional regulator rha061952 from rhodococcus sp. rha1
44 d2o9aa1
not modelled
5.4
10
Fold: Profilin-likeSuperfamily: GAF domain-likeFamily: IclR ligand-binding domain-like45 d1cmca_
not modelled
5.4
50
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Met repressor, MetJ (MetR)