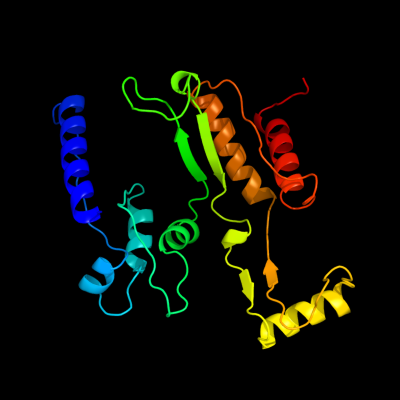
| 1 | d1pb1a_
|
|
 |
100.0 |
100 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
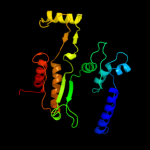
| 2 | c2d4vD_
|
|
 |
100.0 |
69 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
|
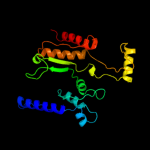
| 3 | d1hqsa_
|
|
 |
100.0 |
71 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 4 | c2e0cA_
|
|
 |
100.0 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:409aa long hypothetical nadp-dependent isocitrate
PDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
|
| 5 | c2d1cB_
|
|
 |
100.0 |
39 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
| 6 | c1tyoA_
|
|
 |
100.0 |
54 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
| 7 | d1w0da_
|
|
 |
100.0 |
31 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 8 | c3fmxX_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas2 putida complexed with nadh
|
| 9 | d1cm7a_
|
|
 |
100.0 |
23 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 10 | d1v53a1
|
|
 |
100.0 |
25 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 11 | d1vlca_
|
|
 |
100.0 |
27 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 12 | d1cnza_
|
|
 |
100.0 |
22 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 13 | d1g2ua_
|
|
 |
100.0 |
27 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 14 | c1zorB_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
| 15 | c3r8wC_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
| 16 | d1xaca_
|
|
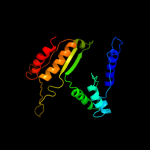 |
100.0 |
27 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 17 | d1wpwa_
|
|
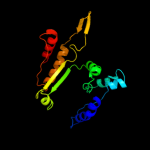 |
100.0 |
29 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 18 | c1x0lB_
|
|
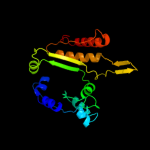 |
100.0 |
30 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
| 19 | c3u1hA_
|
|
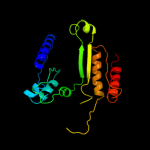 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
|
| 20 | d1a05a_
|
|
 |
100.0 |
25 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 21 | c2uxqB_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase native;
PDBTitle: isocitrate dehydrogenase from the psychrophilic bacterium2 desulfotalea psychrophila: biochemical properties and3 crystal structure analysis
|
| 22 | c3blxL_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:isocitrate dehydrogenase [nad] subunit 2;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 23 | c2qfyE_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
|
| 24 | c3us8A_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
|
| 25 | d1lwda_ |
|
not modelled |
100.0 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 26 | d1t0la_ |
|
not modelled |
100.0 |
12 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
| 27 | c3blxM_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
| 28 | c2iv0A_ |
|
not modelled |
100.0 |
62 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
| 29 | d1hioa_ |
|
not modelled |
20.3 |
17 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 30 | d1f66c_ |
|
not modelled |
17.1 |
14 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 31 | c1f66C_ |
|
not modelled |
17.1 |
14 |
PDB header:structural protein/dna
Chain: C: PDB Molecule:histone h2a.z;
PDBTitle: 2.6 a crystal structure of a nucleosome core particle2 containing the variant histone h2a.z
|
| 32 | d1tzya_ |
|
not modelled |
16.8 |
12 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 33 | c2f8nK_ |
|
not modelled |
16.8 |
13 |
PDB header:structural protein/dna
Chain: K: PDB Molecule:histone h2a type 1;
PDBTitle: 2.9 angstrom x-ray structure of hybrid macroh2a nucleosomes
|
| 34 | d1n1ja_ |
|
not modelled |
16.5 |
10 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 35 | d2pbea1 |
|
not modelled |
15.9 |
5 |
Fold:PAP/OAS1 substrate-binding domain
Superfamily:PAP/OAS1 substrate-binding domain
Family:AadK C-terminal domain-like |
| 36 | d2ffea1 |
|
not modelled |
15.9 |
17 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
| 37 | d2jssa1 |
|
not modelled |
14.5 |
14 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 38 | d1aoic_ |
|
not modelled |
13.6 |
12 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 39 | d1n1jb_ |
|
not modelled |
13.5 |
11 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 40 | d1u35c1 |
|
not modelled |
13.1 |
17 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 41 | d1q77a_ |
|
not modelled |
13.0 |
4 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 42 | c2dumD_ |
|
not modelled |
12.9 |
28 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:hypothetical protein ph0823;
PDBTitle: crystal structure of hypothetical protein, ph0823
|
| 43 | c2p0yA_ |
|
not modelled |
11.8 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein lp_0780;
PDBTitle: crystal structure of q88yi3_lacpl from lactobacillus2 plantarum. northeast structural genomics consortium target3 lpr6
|
| 44 | c1yy3A_ |
|
not modelled |
10.7 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:s-adenosylmethionine:trna ribosyltransferase-
PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
|
| 45 | d1kx3c_ |
|
not modelled |
10.6 |
13 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 46 | d1a7wa_ |
|
not modelled |
10.6 |
13 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Archaeal histone |
| 47 | c3hgmD_ |
|
not modelled |
10.3 |
4 |
PDB header:signaling protein
Chain: D: PDB Molecule:universal stress protein tead;
PDBTitle: universal stress protein tead from the trap transporter2 teaabc of halomonas elongata
|
| 48 | c1v8cA_ |
|
not modelled |
10.2 |
27 |
PDB header:protein binding
Chain: A: PDB Molecule:moad related protein;
PDBTitle: crystal structure of moad related protein from thermus2 thermophilus hb8
|
| 49 | d1ku5a_ |
|
not modelled |
10.1 |
20 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Archaeal histone |
| 50 | c3dwmA_ |
|
not modelled |
9.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:9.5 kda culture filtrate antigen cfp10a;
PDBTitle: crystal structure of mycobacterium tuberculosis cyso, an antigen
|
| 51 | c2jaxA_ |
|
not modelled |
9.3 |
32 |
PDB header:protein binding
Chain: A: PDB Molecule:hypothetical protein tb31.7;
PDBTitle: universal stress protein rv2623 from mycobaterium2 tuberculosis
|
| 52 | d1zcza1 |
|
not modelled |
9.1 |
18 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 53 | d1htaa_ |
|
not modelled |
8.7 |
13 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Archaeal histone |
| 54 | d1k6ka_ |
|
not modelled |
8.4 |
19 |
Fold:Double Clp-N motif
Superfamily:Double Clp-N motif
Family:Double Clp-N motif |
| 55 | d1s32d_ |
|
not modelled |
8.4 |
24 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 56 | d1i7na1 |
|
not modelled |
8.3 |
14 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Synapsin domain |
| 57 | d2bykb1 |
|
not modelled |
8.1 |
10 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:TBP-associated factors, TAFs |
| 58 | c2bykB_ |
|
not modelled |
8.1 |
10 |
PDB header:dna-binding protein
Chain: B: PDB Molecule:chrac-14;
PDBTitle: histone fold heterodimer of the chromatin accessibility2 complex
|
| 59 | c3qd7X_ |
|
not modelled |
8.1 |
22 |
PDB header:hydrolase
Chain: X: PDB Molecule:uncharacterized protein ydal;
PDBTitle: crystal structure of ydal, a stand-alone small muts-related protein2 from escherichia coli
|
| 60 | d1tzyb_ |
|
not modelled |
8.0 |
29 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 61 | c2pbeA_ |
|
not modelled |
7.7 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:aminoglycoside 6-adenylyltransferase;
PDBTitle: crystal structure of an aminoglycoside 6-adenyltransferase2 from bacillus subtilis
|
| 62 | d1kx5c_ |
|
not modelled |
7.7 |
12 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 63 | d1eqzb_ |
|
not modelled |
7.5 |
24 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 64 | d2z3va1 |
|
not modelled |
7.4 |
28 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 65 | c2ppvA_ |
|
not modelled |
7.1 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a protein belonging to the upf0052 (se_0549) from2 staphylococcus epidermidis atcc 12228 at 2.00 a resolution
|
| 66 | c2q7xA_ |
|
not modelled |
7.0 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:upf0052 protein sp_1565;
PDBTitle: crystal structure of a putative phospho transferase (sp_1565) from2 streptococcus pneumoniae tigr4 at 2.00 a resolution
|
| 67 | c3dloC_ |
|
not modelled |
6.9 |
18 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:universal stress protein;
PDBTitle: structure of universal stress protein from archaeoglobus fulgidus
|
| 68 | d1id3c_ |
|
not modelled |
6.7 |
10 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 69 | d1kx5d_ |
|
not modelled |
6.7 |
24 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 70 | d2hzba1 |
|
not modelled |
6.6 |
39 |
Fold:CofD-like
Superfamily:CofD-like
Family:CofD-like |
| 71 | d1jmva_ |
|
not modelled |
6.6 |
22 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:Universal stress protein-like |
| 72 | c2jssA_ |
|
not modelled |
6.6 |
13 |
PDB header:chaperone/nuclear protein
Chain: A: PDB Molecule:chimera of histone h2b.1 and histone h2a.z;
PDBTitle: nmr structure of chaperone chz1 complexed with histone2 h2a.z-h2b
|
| 73 | c2pfsA_ |
|
not modelled |
6.3 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:universal stress protein;
PDBTitle: crystal structure of universal stress protein from nitrosomonas2 europaea
|
| 74 | d2z8la1 |
|
not modelled |
6.3 |
18 |
Fold:OB-fold
Superfamily:Bacterial enterotoxins
Family:Superantigen toxins, N-terminal domain |
| 75 | d2huec1 |
|
not modelled |
6.1 |
23 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 76 | d1eqza_ |
|
not modelled |
6.0 |
12 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 77 | d1id3d_ |
|
not modelled |
5.7 |
19 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 78 | c3idfA_ |
|
not modelled |
5.7 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:usp-like protein;
PDBTitle: the crystal structure of a usp-like protein from wolinella2 succinogenes to 2.0a
|
| 79 | c3fh0A_ |
|
not modelled |
5.5 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative universal stress protein kpn_01444;
PDBTitle: crystal structure of putative universal stress protein kpn_01444 -2 atpase
|
| 80 | c1urqA_ |
|
not modelled |
5.5 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:m-tomosyn isoform;
PDBTitle: crystal structure of neuronal q-snares in complex with2 r-snare motif of tomosyn
|
| 81 | c3ca8B_ |
|
not modelled |
5.4 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:protein ydcf;
PDBTitle: crystal structure of escherichia coli ydcf, an s-adenosyl-l-methionine2 utilizing enzyme
|
| 82 | c3fh2A_ |
|
not modelled |
5.3 |
4 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable atp-dependent protease (heat shock protein);
PDBTitle: the crystal structure of the probable atp-dependent protease (heat2 shock protein) from corynebacterium glutamicum
|
| 83 | d2r61a1 |
|
not modelled |
5.2 |
20 |
Fold:OB-fold
Superfamily:Bacterial enterotoxins
Family:Superantigen toxins, N-terminal domain |
| 84 | c3fpvC_ |
|
not modelled |
5.1 |
28 |
PDB header:heme binding protein
Chain: C: PDB Molecule:extracellular haem-binding protein;
PDBTitle: crystal structure of hbps
|
| 85 | d1bev1_ |
|
not modelled |
5.1 |
29 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |