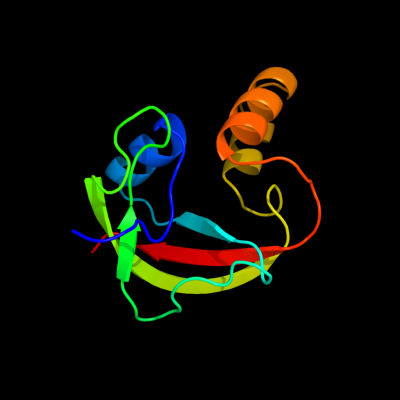
1 d1te7a_
100.0
100
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: yqfB-like2 c3s9xA_
97.0
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: asch domain;PDBTitle: high resolution crystal structure of asch domain from lactobacillus2 crispatus jv v101
3 d1t62a_
96.9
23
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Hypothetical protein EF31334 c3iuwA_
95.0
23
PDB header: rna binding proteinChain: A: PDB Molecule: activating signal cointegrator;PDBTitle: crystal structure of activating signal cointegrator (np_814290.1) from2 enterococcus faecalis v583 at 1.58 a resolution
5 d1xnea_
93.7
18
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain6 d1s04a_
87.9
22
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ProFAR isomerase associated domain7 c2e6zA_
40.7
27
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor spt5;PDBTitle: solution structure of the second kow motif of human2 transcription elongation factor spt5
8 c2dbkA_
34.0
24
PDB header: signaling proteinChain: A: PDB Molecule: crk-like protein;PDBTitle: solution structures of the sh3 domain of human crk-like2 protein
9 d1nxza1
33.5
14
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like10 c2jw4A_
28.6
15
PDB header: signaling proteinChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: nmr solution structure of the n-terminal sh3 domain of2 human nckalpha
11 c1wxtA_
28.2
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein flj21522;PDBTitle: solution structure of the sh3 domain of human hypothetical2 protein flj21522
12 c1vhkA_
24.6
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yqeu;PDBTitle: crystal structure of an hypothetical protein
13 d1o5ua_
23.9
27
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111214 c2i45C_
23.8
9
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of protein nmb1881 from neisseria meningitidis
15 d1wfwa_
23.2
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain16 d1vyva1
23.1
16
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain17 c2xmfA_
22.3
19
PDB header: motor proteinChain: A: PDB Molecule: myosin 1e sh3;PDBTitle: myosin 1e sh3
18 c2kafA_
22.3
31
PDB header: viral protein, rna binding proteinChain: A: PDB Molecule: non-structural protein 3;PDBTitle: solution structure of the sars-unique domain-c from the2 nonstructural protein 3 (nsp3) of the severe acute3 respiratory syndrome coronavirus
19 c3es4B_
22.1
12
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein duf861 with a rmlc-like cupin fold;PDBTitle: crystal structure of protein of unknown function (duf861) with a rmlc-2 like cupin fold (17741406) from agrobacterium tumefaciens str. c583 (dupont) at 1.64 a resolution
20 c3bcwB_
22.0
18
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf861 family protein with a rmlc-like cupin2 fold (bb1179) from bordetella bronchiseptica rb50 at 1.60 a3 resolution
21 c2eyxA_
not modelled
19.8
16
PDB header: signaling proteinChain: A: PDB Molecule: v-crk sarcoma virus ct10 oncogene homologPDBTitle: c-terminal sh3 domain of ct10-regulated kinase
22 c2eqmA_
not modelled
19.3
18
PDB header: transcriptionChain: A: PDB Molecule: phd finger protein 20-like 1;PDBTitle: solution structure of the tudor domain of phd finger2 protein 20-like 1 [homo sapiens]
23 d1j3ta_
not modelled
19.2
15
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain24 d1ep3b1
not modelled
19.2
5
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Riboflavin synthase domain-likeFamily: Ferredoxin reductase FAD-binding domain-like25 d2p5zx2
not modelled
19.1
13
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like26 d1utia_
not modelled
18.1
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain27 d1t3la1
not modelled
17.6
21
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain28 c2ebpA_
not modelled
17.4
19
PDB header: cell adhesionChain: A: PDB Molecule: sam and sh3 domain-containing protein 1;PDBTitle: solution structure of the sh3 domain from human sam and sh32 domain containing protein 1
29 c1zx6A_
not modelled
17.4
12
PDB header: protein bindingChain: A: PDB Molecule: ypr154wp;PDBTitle: high-resolution crystal structure of yeast pin3 sh3 domain
30 d1ng2a1
not modelled
17.3
32
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain31 c2kgtA_
not modelled
16.8
35
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase 6;PDBTitle: solution structure of sh3 domain of ptk6
32 c2cudA_
not modelled
16.8
30
PDB header: signaling proteinChain: A: PDB Molecule: src-like-adapter;PDBTitle: solution structure of the sh3 domain of the human src-like2 adopter protein (slap)
33 c3myxA_
not modelled
16.7
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein pspto_0244;PDBTitle: crystal structure of a pspto_0244 (protein with unknown function which2 belongs to pfam duf861 family) from pseudomonas syringae pv. tomato3 str. dc3000 at 1.30 a resolution
34 c2ct3A_
not modelled
16.5
36
PDB header: signaling proteinChain: A: PDB Molecule: vinexin;PDBTitle: solution structure of the sh3 domain of the vinexin protein
35 c2csqA_
not modelled
16.4
32
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: rim binding protein 2;PDBTitle: solution structure of the second sh3 domain of human rim-2 binding protein 2
36 d1t0ha_
not modelled
16.3
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain37 d1wlpb2
not modelled
16.2
31
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain38 d1udla_
not modelled
15.3
23
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain39 d1ov3a1
not modelled
15.2
32
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain40 c1t0jA_
not modelled
15.1
20
PDB header: signaling proteinChain: A: PDB Molecule: voltage-gated calcium channel subunit beta2a;PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
41 d1ckaa_
not modelled
14.5
32
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain42 c2dl4A_
not modelled
14.3
8
PDB header: signaling proteinChain: A: PDB Molecule: protein stac;PDBTitle: solution structure of the first sh3 domain of stac protein
43 c2js0A_
not modelled
14.3
24
PDB header: signaling proteinChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: solution structure of second sh3 domain of adaptor nck
44 c2eczA_
not modelled
14.1
19
PDB header: signaling proteinChain: A: PDB Molecule: sorbin and sh3 domain-containing protein 1;PDBTitle: solution structure of the sh3 domain of sorbin and sh32 domain-containing protein 1
45 c2krcA_
not modelled
13.9
26
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase subunit delta;PDBTitle: solution structure of the n-terminal domain of bacillus2 subtilis delta subunit of rna polymerase
46 c3kw2A_
not modelled
13.7
19
PDB header: transferaseChain: A: PDB Molecule: probable r-rna methyltransferase;PDBTitle: crystal structure of probable rrna-methyltransferase from2 porphyromonas gingivalis
47 d1jo8a_
not modelled
13.7
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain48 c2ke9A_
not modelled
13.4
29
PDB header: protein bindingChain: A: PDB Molecule: caskin-2;PDBTitle: nmr solution structure of the caskin sh3 domain
49 c1zbe1_
not modelled
13.2
26
PDB header: virusChain: 1: PDB Molecule: coat protein vp1;PDBTitle: foot-and mouth disease virus serotype a1061
50 d2j01e1
not modelled
13.1
24
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L351 d1qqp1_
not modelled
13.1
36
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Picornaviridae-like VP (VP1, VP2, VP3 and VP4)52 d1wlfa1
not modelled
13.0
10
Fold: Cdc48 domain 2-likeSuperfamily: Cdc48 domain 2-likeFamily: Cdc48 domain 2-like53 c2df6A_
not modelled
13.0
38
PDB header: signaling proteinChain: A: PDB Molecule: rho guanine nucleotide exchange factor 7;PDBTitle: crystal structure of the sh3 domain of betapix in complex2 with a high affinity peptide from pak2
54 d1ue9a_
not modelled
13.0
8
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain55 c2bzxA_
not modelled
12.9
25
PDB header: sh3 domainChain: A: PDB Molecule: crk-like protein;PDBTitle: atomic model of crkl-sh3c monomer
56 c1bf5A_
not modelled
12.4
34
PDB header: gene regulation/dnaChain: A: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: tyrosine phosphorylated stat-1/dna complex
57 d1sema_
not modelled
12.4
28
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain58 c2jxbA_
not modelled
12.3
11
PDB header: signaling protein complexChain: A: PDB Molecule: t-cell surface glycoprotein cd3 epsilon chain,PDBTitle: structure of cd3epsilon-nck2 first sh3 domain complex
59 c2cubA_
not modelled
12.1
24
PDB header: signaling proteinChain: A: PDB Molecule: cytoplasmic protein nck1;PDBTitle: solution structure of the sh3 domain of the human2 cytoplasmic protein nck1
60 c2krnA_
not modelled
12.1
28
PDB header: signaling proteinChain: A: PDB Molecule: cd2-associated protein;PDBTitle: high resolution structure of the second sh3 domain of cd2ap
61 c1g5vA_
not modelled
11.9
26
PDB header: translationChain: A: PDB Molecule: survival motor neuron protein 1;PDBTitle: solution structure of the tudor domain of the human smn2 protein
62 c3cgnA_
not modelled
11.9
26
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of thermophilic slyd
63 d1i07a_
not modelled
11.8
22
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain64 c2yuoA_
not modelled
11.7
20
PDB header: signaling proteinChain: A: PDB Molecule: run and tbc1 domain containing 3;PDBTitle: solution structure of the sh3 domain of mouse run and tbc12 domain containing 3
65 d1h8ka_
not modelled
11.6
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain66 c2o2oA_
not modelled
11.5
24
PDB header: protein bindingChain: A: PDB Molecule: sh3-domain kinase-binding protein 1;PDBTitle: solution structure of domain b from human cin85 protein
67 c2egcA_
not modelled
11.4
26
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structure of the fifth sh3 domain from human2 kiaa0418 protein
68 d1efna_
not modelled
11.4
12
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain69 d1uhfa_
not modelled
11.4
11
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain70 d1pwta_
not modelled
11.4
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain71 d1yfua1
not modelled
11.4
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: 3-hydroxyanthranilic acid dioxygenase-like72 c2dx1A_
not modelled
11.3
30
PDB header: signaling proteinChain: A: PDB Molecule: rho guanine nucleotide exchange factor 4;PDBTitle: crystal structure of rhogef protein asef
73 d2pyta1
not modelled
11.1
24
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like74 c2pqhA_
not modelled
11.0
20
PDB header: structural proteinChain: A: PDB Molecule: spectrin alpha chain, brain;PDBTitle: structure of sh3 chimera with a type ii ligand linked to the chain c-2 terminal
75 c1zlmA_
not modelled
10.9
23
PDB header: signaling proteinChain: A: PDB Molecule: osteoclast stimulating factor 1;PDBTitle: crystal structure of the sh3 domain of human osteoclast2 stimulating factor
76 d2dp9a1
not modelled
10.9
16
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Hypothetical protein TTHA011377 c2drmB_
not modelled
10.8
28
PDB header: contractile proteinChain: B: PDB Molecule: acanthamoeba myosin ib;PDBTitle: acanthamoeba myosin i sh3 domain bound to acan125
78 c2egaA_
not modelled
10.7
23
PDB header: signaling proteinChain: A: PDB Molecule: sh3 and px domain-containing protein 2a;PDBTitle: solution structure of the first sh3 domain from human2 kiaa0418 protein
79 d1mhna_
not modelled
10.6
17
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: Tudor domain80 c1yn8E_
not modelled
10.6
16
PDB header: unknown functionChain: E: PDB Molecule: nap1-binding protein 2;PDBTitle: sh3 domain of yeast nbp2
81 d1lcka1
not modelled
10.6
15
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain82 d1vyua1
not modelled
10.5
17
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain83 c1w1fA_
not modelled
10.5
15
PDB header: sh3-domainChain: A: PDB Molecule: tyrosine-protein kinase lyn;PDBTitle: sh3 domain of human lyn tyrosine kinase
84 d1gcqa_
not modelled
10.4
23
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain85 c1wxbA_
not modelled
10.4
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: epidermal growth factor receptor pathwayPDBTitle: solution structure of the sh3 domain from human epidermal2 growth factor receptor pathway substrate 8-like protein
86 c2l0aA_
not modelled
10.4
24
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 1;PDBTitle: solution nmr structure of signal transducing adapter molecule 1 stam-12 from homo sapiens, northeast structural genomics consortium target3 hr4479e
87 c1x2qA_
not modelled
10.3
23
PDB header: signaling proteinChain: A: PDB Molecule: signal transducing adapter molecule 2;PDBTitle: solution structure of the sh3 domain of the signal2 transducing adaptor molecule 2
88 d2gycb1
not modelled
10.3
14
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L389 c2jmcA_
not modelled
10.2
20
PDB header: signaling proteinChain: A: PDB Molecule: spectrin alpha chain, brain and p41 peptidePDBTitle: chimer between spc-sh3 and p41
90 d1awwa_
not modelled
10.1
8
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain91 d1h5ba_
not modelled
10.1
36
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)92 c3rebD_
not modelled
10.0
15
PDB header: protein bindingChain: D: PDB Molecule: tyrosine-protein kinase hck;PDBTitle: hiv-1 nef protein in complex with engineered hck-sh3 domain
93 d1y57a1
not modelled
9.9
24
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain94 c2oi3A_
not modelled
9.8
15
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase hck;PDBTitle: nmr structure analysis of the hematopoetic cell kinase sh32 domain complexed with an artificial high affinity ligand3 (pd1)
95 d2zjrb1
not modelled
9.7
10
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: Ribosomal protein L396 c2hdeA_
not modelled
9.6
25
PDB header: transcriptionChain: A: PDB Molecule: histone deacetylase complex subunit sap18;PDBTitle: solution structure of human sap18
97 c3h8uA_
not modelled
9.6
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized conserved protein with double-strandedPDBTitle: crystal structure of uncharacterized conserved protein with double-2 stranded beta-helix domain (yp_001338853.1) from klebsiella3 pneumoniae subsp. pneumoniae mgh 78578 at 1.80 a resolution
98 d1e6ga_
not modelled
9.6
20
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain99 c3nmzD_
not modelled
9.6
32
PDB header: cell adhesion/cell cycleChain: D: PDB Molecule: rho guanine nucleotide exchange factor 4;PDBTitle: crytal structure of apc complexed with asef