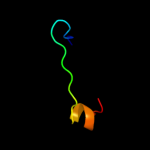
1 d1x3za1
47.4
9
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core2 c3mswA_
37.2
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (bf3112) from2 bacteroides fragilis nctc 9343 at 1.90 a resolution
3 d2f4ma1
36.0
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Transglutaminase core4 c3eswA_
29.7
13
PDB header: hydrolaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-glucosaminyl)asparaginePDBTitle: complex of yeast pngase with glcnac2-iac.
5 c1j5qB_
21.8
24
PDB header: viral proteinChain: B: PDB Molecule: major capsid protein;PDBTitle: the structure and evolution of the major capsid protein of a large,2 lipid-containing, dna virus.
6 c2x12A_
21.2
27
PDB header: cell adhesionChain: A: PDB Molecule: fimbriae-associated protein fap1;PDBTitle: ph-induced modulation of streptococcus parasanguinis2 adhesion by fap1 fimbriae
7 d1pk6c_
21.1
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like8 d1ei5a1
19.4
32
Fold: Streptavidin-likeSuperfamily: D-aminopeptidase, middle and C-terminal domainsFamily: D-aminopeptidase, middle and C-terminal domains9 d1pk6b_
18.3
27
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like10 c2oy7A_
17.6
11
PDB header: membrane proteinChain: A: PDB Molecule: outer surface protein a;PDBTitle: the crystal structure of ospa mutant
11 d1bcoa1
17.5
11
Fold: mu transposase, C-terminal domainSuperfamily: mu transposase, C-terminal domainFamily: mu transposase, C-terminal domain12 d1c3ha_
17.2
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like13 c1gr3A_
14.6
36
PDB header: collagenChain: A: PDB Molecule: collagen x;PDBTitle: structure of the human collagen x nc1 trimer
14 d1gr3a_
14.6
36
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like15 c3kifD_
14.0
32
PDB header: sugar binding proteinChain: D: PDB Molecule: 5-bladed -propeller lectin;PDBTitle: the crystal structures of two fragments truncated from 5-bladed -2 propeller lectin, tachylectin-2 (lib1-b7-18 and lib2-d2-15)
16 c2ka3C_
12.2
38
PDB header: structural proteinChain: C: PDB Molecule: emilin-1;PDBTitle: structure of emilin-1 c1q-like domain
17 c7apiB_
11.8
6
PDB header: proteinase inhibitorChain: B: PDB Molecule: alpha 1-antitrypsin;PDBTitle: the s variant of human alpha1-antitrypsin, structure and implications2 for function and metabolism
18 d2jdqd1
11.1
17
Fold: PB2 C-terminal domain-likeSuperfamily: PB2 C-terminal domain-likeFamily: PB2 C-terminal domain-like19 d1pk6a_
11.1
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like20 c2vtwF_
11.0
27
PDB header: viral proteinChain: F: PDB Molecule: fiber protein 2;PDBTitle: structure of the c-terminal head domain of the fowl2 adenovirus type 1 short fibre
21 d1f1sa3
not modelled
10.8
23
Fold: Hyaluronate lyase-like, C-terminal domainSuperfamily: Hyaluronate lyase-like, C-terminal domainFamily: Hyaluronate lyase-like, C-terminal domain22 c1jjoE_
not modelled
10.5
22
PDB header: signaling proteinChain: E: PDB Molecule: neuroserpin;PDBTitle: crystal structure of mouse neuroserpin (cleaved form)
23 d1m3ya2
not modelled
10.4
33
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Group II dsDNA viruses VPFamily: Major capsid protein vp5424 d1o91a_
not modelled
10.4
31
Fold: TNF-likeSuperfamily: TNF-likeFamily: TNF-like25 c1o91B_
not modelled
10.4
31
PDB header: collagenChain: B: PDB Molecule: collagen alpha 1(viii) chain;PDBTitle: crystal structure of a collagen viii nc1 domain trimer
26 c1m4xC_
not modelled
9.7
33
PDB header: virusChain: C: PDB Molecule: pbcv-1 virus capsid;PDBTitle: pbcv-1 virus capsid, quasi-atomic model
27 d1w9pa2
not modelled
9.6
18
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain28 c1hleB_
not modelled
9.5
18
PDB header: hydrolase inhibitor(serine proteinase)Chain: B: PDB Molecule: horse leukocyte elastase inhibitor;PDBTitle: crystal structure of cleaved equine leucocyte elastase2 inhibitor determined at 1.95 angstroms resolution
29 d1brwa3
not modelled
9.4
23
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain30 d2tpta3
not modelled
9.0
19
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain31 d1ll7a2
not modelled
8.9
23
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain32 c3f02C_
not modelled
8.8
16
PDB header: hydrolase inhibitorChain: C: PDB Molecule: neuroserpin;PDBTitle: cleaved human neuroserpin
33 d1ulva3
not modelled
8.8
42
Fold: Immunoglobulin-like beta-sandwichSuperfamily: CBD9-likeFamily: Glucodextranase, domain C34 c2qshA_
not modelled
8.7
25
PDB header: dna binding protein/dnaChain: A: PDB Molecule: dna repair protein rad4;PDBTitle: crystal structure of rad4-rad23 bound to a mismatch dna
35 d1zaka2
not modelled
8.6
33
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain36 c2iqcA_
not modelled
8.2
25
PDB header: protein bindingChain: A: PDB Molecule: fanconi anemia group f protein;PDBTitle: crystal structure of human fancf protein that functions in2 the assembly of a dna damage signaling complex
37 c3mmlD_
not modelled
8.1
21
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
38 c3rduA_
not modelled
8.1
18
PDB header: biotin binding proteinChain: A: PDB Molecule: streptavidin;PDBTitle: crystal structure of r7-2 streptavidin complexed with peg
39 c2h4qB_
not modelled
8.0
18
PDB header: hydrolase inhibitorChain: B: PDB Molecule: heterochromatin-associated protein ment;PDBTitle: crystal structure of a m-loop deletion variant of ment in2 the cleaved conformation
40 d1zunb2
not modelled
7.8
25
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domainFamily: EF-Tu/eEF-1alpha/eIF2-gamma C-terminal domain41 c1dfcB_
not modelled
7.7
16
PDB header: structural proteinChain: B: PDB Molecule: fascin;PDBTitle: crystal structure of human fascin, an actin-crosslinking protein
42 c9paiB_
not modelled
7.6
12
PDB header: hydrolase inhibitorChain: B: PDB Molecule: protein (plasminogen activator inhibitor-1) residues 365-PDBTitle: cleaved substrate variant of plasminogen activator inhibitor-1
43 c1wwtA_
not modelled
7.5
17
PDB header: ligaseChain: A: PDB Molecule: threonyl-trna synthetase, cytoplasmic;PDBTitle: solution structure of the tgs domain from human threonyl-2 trna synthetase
44 d2ax3a2
not modelled
7.3
18
Fold: YjeF N-terminal domain-likeSuperfamily: YjeF N-terminal domain-likeFamily: YjeF N-terminal domain-like45 d1n7oa2
not modelled
7.3
16
Fold: Hyaluronate lyase-like, C-terminal domainSuperfamily: Hyaluronate lyase-like, C-terminal domainFamily: Hyaluronate lyase-like, C-terminal domain46 c3kzsD_
not modelled
7.1
13
PDB header: hydrolaseChain: D: PDB Molecule: glycosyl hydrolase family 5;PDBTitle: crystal structure of glycosyl hydrolase family 5 (np_809925.1) from2 bacteroides thetaiotaomicron vpi-5482 at 2.10 a resolution
47 c2rivB_
not modelled
7.0
6
PDB header: signaling proteinChain: B: PDB Molecule: thyroxine-binding globulin;PDBTitle: crystal structure of the reactive loop cleaved human thyroxine binding2 globulin
48 d1azpa_
not modelled
6.9
25
Fold: SH3-like barrelSuperfamily: Chromo domain-likeFamily: "Histone-like" proteins from archaea49 d1k8wa3
not modelled
6.8
27
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain50 d1gxma_
not modelled
6.8
33
Fold: alpha/alpha toroidSuperfamily: Family 10 polysaccharide lyaseFamily: Family 10 polysaccharide lyase51 d1vpra1
not modelled
6.8
60
Fold: LipocalinsSuperfamily: LipocalinsFamily: Dinoflagellate luciferase repeat52 c2l92A_
not modelled
6.7
25
PDB header: dna binding proteinChain: A: PDB Molecule: histone family protein nucleoid-structuring protein h-ns;PDBTitle: solution structure of the c-terminal domain of h-ns like protein bv3f
53 d1gkpa1
not modelled
6.5
15
Fold: Composite domain of metallo-dependent hydrolasesSuperfamily: Composite domain of metallo-dependent hydrolasesFamily: Hydantoinase (dihydropyrimidinase)54 c2nqoB_
not modelled
6.4
12
PDB header: transferaseChain: B: PDB Molecule: gamma-glutamyltranspeptidase;PDBTitle: crystal structure of helicobacter pylori gamma-glutamyltranspeptidase
55 c2ksnA_
not modelled
6.3
22
PDB header: signaling proteinChain: A: PDB Molecule: ubiquitin domain-containing protein 2;PDBTitle: solution structure of the n-terminal domain of dc-ubp/ubtd2
56 c1pgsA_
not modelled
6.3
63
PDB header: endoglycosidaseChain: A: PDB Molecule: peptide-n(4)-(n-acetyl-beta-d-glucosaminyl)PDBTitle: the three-dimensional structure of pngase f, a2 glycosylasparaginase from flavobacterium meningosepticum
57 d1w07a2
not modelled
5.9
13
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains58 c2uywA_
not modelled
5.8
18
PDB header: glycoproteinChain: A: PDB Molecule: xenavidin;PDBTitle: crystal structure of xenavidin
59 c3iyoA_
not modelled
5.7
15
PDB header: virusChain: A: PDB Molecule: capsid protein;PDBTitle: cryo-em model of virion-sized hev virion-sized capsid
60 c2dafA_
not modelled
5.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: flj35834 protein;PDBTitle: solution structure of the novel identified ubiquitin-like2 domain in the human hypothetical protein flj35834
61 c2yh5A_
not modelled
5.7
3
PDB header: lipid binding proteinChain: A: PDB Molecule: dapx protein;PDBTitle: structure of the c-terminal domain of bamc
62 c2elpA_
not modelled
5.6
26
PDB header: transcriptionChain: A: PDB Molecule: zinc finger protein 406;PDBTitle: solution structure of the 13th c2h2 zinc finger of human2 zinc finger protein 406
63 c3nqhA_
not modelled
5.6
20
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase;PDBTitle: crystal structure of a glycosyl hydrolase (bt_2959) from bacteroides2 thetaiotaomicron vpi-5482 at 2.11 a resolution
64 c3cw4A_
not modelled
5.5
17
PDB header: transferaseChain: A: PDB Molecule: polymerase basic protein 2;PDBTitle: large c-terminal domain of influenza a virus rna-dependent polymerase2 pb2
65 d2rh3a1
not modelled
5.5
40
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: VirC2-like66 d1s6la1
not modelled
5.4
44
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MerB N-terminal domain-like67 d1stma_
not modelled
5.4
38
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Satellite virusesFamily: Satellite viruses68 d1z84a1
not modelled
5.3
33
Fold: HIT-likeSuperfamily: HIT-likeFamily: Hexose-1-phosphate uridylyltransferase69 d1goia3
not modelled
5.2
27
Fold: FKBP-likeSuperfamily: Chitinase insertion domainFamily: Chitinase insertion domain70 c1e0aB_
not modelled
5.2
15
PDB header: signalling proteinChain: B: PDB Molecule: serine/threonine-protein kinase pak-alpha;PDBTitle: cdc42 complexed with the gtpase binding domain of p212 activated kinase
71 c3fm2A_
not modelled
5.1
9
PDB header: heme-binding proteinChain: A: PDB Molecule: uncharacterized protein, distantly related to a hemePDBTitle: crystal structure of a putative heme-binding protein (ava_4353) from2 anabaena variabilis atcc 29413 at 1.80 a resolution
72 d2ddha2
not modelled
5.1
17
Fold: Bromodomain-likeSuperfamily: Acyl-CoA dehydrogenase C-terminal domain-likeFamily: acyl-CoA oxidase C-terminal domains73 c3ew2A_
not modelled
5.1
14
PDB header: unknown functionChain: A: PDB Molecule: rhizavidin;PDBTitle: crystal structure of rhizavidin-biotin complex
74 c2c1sA_
not modelled
5.1
9
PDB header: biotin binding proteinChain: A: PDB Molecule: biotin binding protein a;PDBTitle: x-ray structure of biotin binding protein from chicken
75 d1es0a2
not modelled
5.0
29
Fold: MHC antigen-recognition domainSuperfamily: MHC antigen-recognition domainFamily: MHC antigen-recognition domain