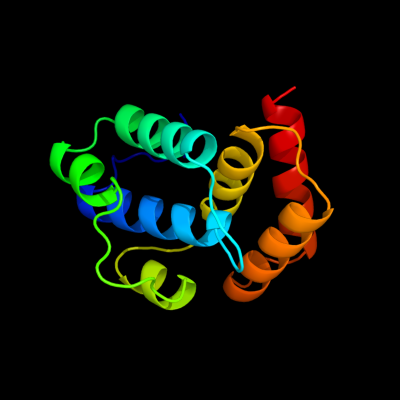
1 d2ou3a1
99.6
17
Fold: TerB-likeSuperfamily: TerB-likeFamily: COG3793-like2 c2jxuA_
98.7
9
PDB header: unknown functionChain: A: PDB Molecule: terb;PDBTitle: nmr solution structure of kp-terb, a tellurite resistance2 protein from klebsiella pneumoniae
3 d1z67a1
86.5
18
Fold: YidB-likeSuperfamily: YidB-likeFamily: YidB-like4 c3hugJ_
56.3
23
PDB header: transcription/membrane proteinChain: J: PDB Molecule: probable conserved membrane protein;PDBTitle: crystal structure of mycobacterium tuberculosis anti-sigma factor rsla2 in complex with -35 promoter binding domain of sigl
5 d1aisb2
56.1
8
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain6 d1ln4a_
47.6
30
Fold: IF3-likeSuperfamily: YhbY-likeFamily: YhbY-like7 d2p7vb1
42.1
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain8 d1yzma1
39.3
25
Fold: Long alpha-hairpinSuperfamily: Rabenosyn-5 Rab-binding domain-likeFamily: Rabenosyn-5 Rab-binding domain-like9 d1z0kb1
38.9
25
Fold: Long alpha-hairpinSuperfamily: Rabenosyn-5 Rab-binding domain-likeFamily: Rabenosyn-5 Rab-binding domain-like10 d1ku7a_
37.5
18
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain11 c3fymA_
36.9
7
PDB header: dna binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: the 1a structure of ymfm, a putative dna-binding membrane2 protein from staphylococcus aureus
12 c3ol4B_
35.3
6
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
13 d1rq8a_
34.9
25
Fold: IF3-likeSuperfamily: YhbY-likeFamily: YhbY-like14 c2iv1J_
34.4
10
PDB header: lyaseChain: J: PDB Molecule: cyanate hydratase;PDBTitle: site directed mutagenesis of key residues involved in the2 catalytic mechanism of cyanase
15 d1ttya_
33.9
24
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain16 d1z0jb1
33.4
42
Fold: Long alpha-hairpinSuperfamily: Rabenosyn-5 Rab-binding domain-likeFamily: Rabenosyn-5 Rab-binding domain-like17 d1jo0a_
33.2
25
Fold: IF3-likeSuperfamily: YhbY-likeFamily: YhbY-like18 c3obkH_
32.9
20
PDB header: lyaseChain: H: PDB Molecule: delta-aminolevulinic acid dehydratase;PDBTitle: crystal structure of delta-aminolevulinic acid dehydratase2 (porphobilinogen synthase) from toxoplasma gondii me49 in complex3 with the reaction product porphobilinogen
19 d1ctda_
32.4
33
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like20 d1vkeb_
32.4
11
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: CMD-like21 c2l27B_
not modelled
32.2
12
PDB header: membrane protein, peptide binding proteiChain: B: PDB Molecule: peptide agonist;PDBTitle: nmr structure of the ecd1 of crf-r1 in complex with a peptide agonist
22 c2rmfA_
not modelled
30.6
21
PDB header: hormoneChain: A: PDB Molecule: urocortin;PDBTitle: human urocortin 1
23 c3m3hA_
not modelled
30.6
20
PDB header: transferaseChain: A: PDB Molecule: orotate phosphoribosyltransferase;PDBTitle: 1.75 angstrom resolution crystal structure of an orotate2 phosphoribosyltransferase from bacillus anthracis str. 'ames3 ancestor'
24 d1msza_
not modelled
30.5
6
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain25 c1mszA_
not modelled
30.5
6
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein smubp-2;PDBTitle: solution structure of the r3h domain from human smubp-2
26 c2wusR_
not modelled
30.5
9
PDB header: structural proteinChain: R: PDB Molecule: putative uncharacterized protein;PDBTitle: bacterial actin mreb assembles in complex with cell shape2 protein rodz
27 d1dwka1
not modelled
26.2
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Cyanase N-terminal domain28 d1vola2
not modelled
24.6
11
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain29 d2gyqa1
not modelled
23.9
16
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: YciF-like30 d1d5ya1
not modelled
22.5
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator31 c3t72o_
not modelled
22.4
18
PDB header: transcription/dnaChain: O: PDB Molecule: pho box dna (strand 1);PDBTitle: phob(e)-sigma70(4)-(rnap-betha-flap-tip-helix)-dna transcription2 activation sub-complex
32 c2k29A_
not modelled
22.2
26
PDB header: transcriptionChain: A: PDB Molecule: antitoxin relb;PDBTitle: structure of the dbd domain of e. coli antitoxin relb
33 c3oi8B_
not modelled
21.8
22
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: the crystal structure of functionally unknown conserved protein domain2 from neisseria meningitidis mc58
34 d1ku3a_
not modelled
20.4
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain35 d1ufma_
not modelled
19.4
27
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PCI domain (PINT motif)36 c2k9lA_
not modelled
18.5
8
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: structure of the core binding domain of sigma54
37 c3beyC_
not modelled
18.1
19
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: conserved protein o27018;PDBTitle: crystal structure of the protein o27018 from methanobacterium2 thermoautotrophicum. northeast structural genomics consortium target3 tt217
38 c2a6eF_
not modelled
17.8
18
PDB header: transferaseChain: F: PDB Molecule: rna polymerase sigma factor rpod;PDBTitle: crystal structure of the t. thermophilus rna polymerase2 holoenzyme
39 c1p8cD_
not modelled
17.7
10
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of tm1620 (apc4843) from thermotoga2 maritima
40 c3hjlA_
not modelled
16.1
11
PDB header: proton transportChain: A: PDB Molecule: flagellar motor switch protein flig;PDBTitle: the structure of full-length flig from aquifex aeolicus
41 d2gtia2
not modelled
16.0
9
Fold: EndoU-likeSuperfamily: EndoU-likeFamily: Nsp15 C-terminal domain-like42 c2zetD_
not modelled
15.2
10
PDB header: signaling proteinChain: D: PDB Molecule: melanophilin;PDBTitle: crystal structure of the small gtpase rab27b complexed with2 the slp homology domain of slac2-a/melanophilin
43 d1hqva_
not modelled
15.1
11
Fold: EF Hand-likeSuperfamily: EF-handFamily: Penta-EF-hand proteins44 c2i7aA_
not modelled
14.5
13
PDB header: hydrolaseChain: A: PDB Molecule: calpain 13;PDBTitle: domain iv of human calpain 13
45 d1l6sa_
not modelled
14.3
23
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)46 d1vola1
not modelled
13.6
6
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Transcription factor IIB (TFIIB), core domain47 d1bl0a1
not modelled
13.1
15
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: AraC type transcriptional activator48 c1l9uH_
not modelled
13.1
24
PDB header: transcriptionChain: H: PDB Molecule: sigma factor siga;PDBTitle: thermus aquaticus rna polymerase holoenzyme at 4 a2 resolution
49 d2c1ha1
not modelled
12.9
29
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)50 d1w8ia_
not modelled
12.7
37
Fold: PIN domain-likeSuperfamily: PIN domain-likeFamily: PIN domain51 c1y66D_
not modelled
12.4
20
PDB header: de novo proteinChain: D: PDB Molecule: engrailed homeodomain;PDBTitle: dioxane contributes to the altered conformation and2 oligomerization state of a designed engrailed homeodomain3 variant
52 c2rhbD_
not modelled
11.7
27
PDB header: viral proteinChain: D: PDB Molecule: uridylate-specific endoribonuclease;PDBTitle: crystal structure of nsp15-h234a mutant- hexamer in2 asymmetric unit
53 d1h7na_
not modelled
11.5
29
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)54 d1ojra_
not modelled
11.3
13
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase55 c2ib1A_
not modelled
11.0
11
PDB header: apoptosisChain: A: PDB Molecule: death domain containing membrane protein nradd;PDBTitle: solution structure of p45 death domain
56 d2e74g1
not modelled
11.0
20
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex57 d2q0ta1
not modelled
10.9
23
Fold: AhpD-likeSuperfamily: AhpD-likeFamily: AhpD58 d1lmb3_
not modelled
10.7
16
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors59 d1bi5a2
not modelled
10.7
15
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like60 d2ozka2
not modelled
10.6
27
Fold: EndoU-likeSuperfamily: EndoU-likeFamily: Nsp15 C-terminal domain-like61 c1vf5G_
not modelled
10.5
20
PDB header: photosynthesisChain: G: PDB Molecule: protein pet g;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
62 d1vf5g_
not modelled
10.5
20
Fold: Single transmembrane helixSuperfamily: PetG subunit of the cytochrome b6f complexFamily: PetG subunit of the cytochrome b6f complex63 d2cyya1
not modelled
10.4
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain64 d1mkma1
not modelled
10.2
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Transcriptional regulator IclR, N-terminal domain65 c2lfcA_
not modelled
9.9
13
PDB header: oxidoreductaseChain: A: PDB Molecule: fumarate reductase, flavoprotein subunit;PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
66 c3bh1A_
not modelled
9.8
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0371 protein dip2346;PDBTitle: crystal structure of protein dip2346 from corynebacterium diphtheriae
67 d2h85a2
not modelled
9.8
27
Fold: EndoU-likeSuperfamily: EndoU-likeFamily: Nsp15 C-terminal domain-like68 d2hf5a1
not modelled
9.8
29
Fold: EF Hand-likeSuperfamily: EF-handFamily: Calmodulin-like69 d1ngra_
not modelled
9.6
15
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD70 c2ev2B_
not modelled
9.6
15
PDB header: lyaseChain: B: PDB Molecule: hypothetical protein rv1264/mt1302;PDBTitle: structure of rv1264n, the regulatory domain of the mycobacterial2 adenylyl cylcase rv1264, at ph 8.5
71 d2p90a1
not modelled
9.6
21
Fold: Phosphorylase/hydrolase-likeSuperfamily: Cgl1923-likeFamily: Cgl1923-like72 c2of5K_
not modelled
9.5
15
PDB header: apoptosisChain: K: PDB Molecule: leucine-rich repeat and death domain-containingPDBTitle: oligomeric death domain complex
73 d1rp3a1
not modelled
9.3
8
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma3 domain74 d2tpta1
not modelled
9.2
21
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain75 c1ichA_
not modelled
9.1
15
PDB header: apoptosisChain: A: PDB Molecule: tumor necrosis factor receptor-1;PDBTitle: solution structure of the tumor necrosis factor receptor-12 death domain
76 d1icha_
not modelled
9.1
15
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD77 d1tw3a1
not modelled
8.9
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Plant O-methyltransferase, N-terminal domain78 d2bida_
not modelled
8.8
25
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death79 d1o17a1
not modelled
8.8
19
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain80 c2gtiA_
not modelled
8.5
9
PDB header: viral proteinChain: A: PDB Molecule: replicase polyprotein 1ab;PDBTitle: mutation of mhv coronavirus non-structural protein nsp15 (f307l)
81 d1khda1
not modelled
8.5
9
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain82 c1wxpA_
not modelled
8.2
25
PDB header: transport proteinChain: A: PDB Molecule: tho complex subunit 1;PDBTitle: solution structure of the death domain of nuclear matrix2 protein p84
83 d2ofya1
not modelled
8.2
15
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like84 c2cpmA_
not modelled
8.2
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: sperm-associated antigen 7;PDBTitle: solution structure of the r3h domain of human sperm-2 associated antigen 7
85 c2p90B_
not modelled
8.1
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hypothetical protein cgl1923;PDBTitle: the crystal structure of a protein of unknown function from2 corynebacterium glutamicum atcc 13032
86 c2wvsD_
not modelled
7.9
24
PDB header: hydrolaseChain: D: PDB Molecule: alpha-l-fucosidase;PDBTitle: crystal structure of an alpha-l-fucosidase gh29 trapped2 covalent intermediate from bacteroides thetaiotaomicron in3 complex with 2-fluoro-fucosyl fluoride using an e288q4 mutant
87 c1gpmD_
not modelled
7.9
14
PDB header: transferase (glutamine amidotransferase)Chain: D: PDB Molecule: gmp synthetase;PDBTitle: escherichia coli gmp synthetase complexed with amp and pyrophosphate
88 c2z2sD_
not modelled
7.9
11
PDB header: transcriptionChain: D: PDB Molecule: anti-sigma factor chrr, transcriptional activator chrr;PDBTitle: crystal structure of rhodobacter sphaeroides sige in complex with the2 anti-sigma chrr
89 d2gf5a1
not modelled
7.8
30
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD90 c3omtA_
not modelled
7.8
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: putative antitoxin component, chu_2935 protein, from xre family from2 prevotella buccae.
91 c2vn2B_
not modelled
7.7
15
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
92 d1brwa1
not modelled
7.7
17
Fold: Methionine synthase domain-likeSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase N-terminal domain93 d1or7a1
not modelled
7.7
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Sigma3 and sigma4 domains of RNA polymerase sigma factorsFamily: Sigma4 domain94 d1t07a_
not modelled
7.7
14
Fold: YggX-likeSuperfamily: YggX-likeFamily: YggX-like95 d1u0ua2
not modelled
7.7
20
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like96 c3t98A_
not modelled
7.6
33
PDB header: protein transportChain: A: PDB Molecule: nuclear pore complex protein nup54;PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup54/nup58
97 d1ee0a2
not modelled
7.6
25
Fold: Thiolase-likeSuperfamily: Thiolase-likeFamily: Chalcone synthase-like98 c2yqfA_
not modelled
7.6
15
PDB header: protein bindingChain: A: PDB Molecule: ankyrin-1;PDBTitle: solution structure of the death domain of ankyrin-1
99 c3e35A_
not modelled
7.5
11
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein sco1997;PDBTitle: actinobacteria-specific protein of unknown function, sco1997