| 1 |
|
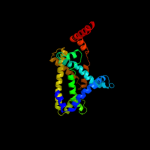
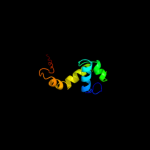
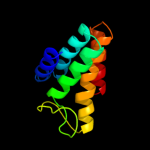
PDB 2knc chain A
Region: 301 - 360
Aligned: 49
Modelled: 60
Confidence: 67.0%
Identity: 27%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 2 |
|
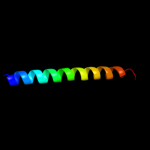
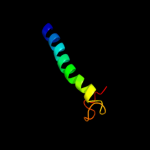
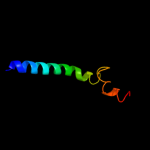
PDB 1kkx chain A
Region: 297 - 364
Aligned: 68
Modelled: 68
Confidence: 37.2%
Identity: 13%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: ARID-like
Family: ARID domain
Phyre2
| 3 |
|
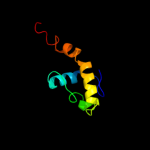
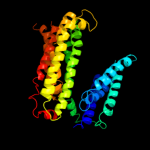
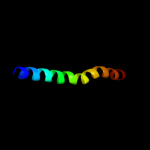
PDB 3eh4 chain A
Region: 318 - 370
Aligned: 53
Modelled: 53
Confidence: 23.3%
Identity: 11%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c oxidase subunit 1;
PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
Phyre2
| 4 |
|
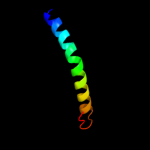
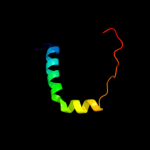
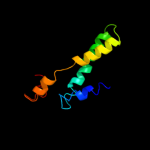
PDB 1xme chain A domain 1
Region: 318 - 370
Aligned: 53
Modelled: 53
Confidence: 19.6%
Identity: 11%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 5 |
|
PDB 2jln chain A
Region: 308 - 365
Aligned: 58
Modelled: 58
Confidence: 10.5%
Identity: 10%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 6 |
|
PDB 2jwa chain A
Region: 305 - 333
Aligned: 29
Modelled: 29
Confidence: 8.8%
Identity: 10%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 68 - 367
Aligned: 244
Modelled: 253
Confidence: 8.2%
Identity: 12%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 2hg5 chain D
Region: 318 - 353
Aligned: 36
Modelled: 36
Confidence: 8.1%
Identity: 8%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 9 |
|
PDB 1ryu chain A
Region: 297 - 365
Aligned: 69
Modelled: 69
Confidence: 7.8%
Identity: 16%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: ARID-like
Family: ARID domain
Phyre2
| 10 |
|
PDB 3qnq chain D
Region: 324 - 360
Aligned: 37
Modelled: 37
Confidence: 7.5%
Identity: 5%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 11 |
|
PDB 2eqy chain A
Region: 297 - 370
Aligned: 74
Modelled: 74
Confidence: 7.4%
Identity: 14%
PDB header:dna binding protein
Chain: A: PDB Molecule:jumonji, at rich interactive domain 1b;
PDBTitle: solution structure of the arid domain of jarid1b protein
Phyre2
| 12 |
|
PDB 2jo1 chain A
Region: 324 - 365
Aligned: 42
Modelled: 42
Confidence: 7.0%
Identity: 14%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 13 |
|
PDB 3mku chain A
Region: 3 - 361
Aligned: 341
Modelled: 341
Confidence: 6.8%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
Phyre2
| 14 |
|
PDB 2iz1 chain C
Region: 329 - 370
Aligned: 42
Modelled: 42
Confidence: 6.8%
Identity: 14%
PDB header:oxidoreductase
Chain: C: PDB Molecule:6-phosphogluconate dehydrogenase, decarboxylating;
PDBTitle: 6pdh complexed with pex inhibitor synchrotron data
Phyre2
| 15 |
|
PDB 2qts chain A
Region: 319 - 341
Aligned: 23
Modelled: 23
Confidence: 6.3%
Identity: 35%
PDB header:membrane protein
Chain: A: PDB Molecule:acid-sensing ion channel;
PDBTitle: structure of an acid-sensing ion channel 1 at 1.9 a resolution and low2 ph
Phyre2
| 16 |
|
PDB 3rlb chain A
Region: 241 - 346
Aligned: 97
Modelled: 106
Confidence: 6.2%
Identity: 19%
PDB header:thiamine-binding protein
Chain: A: PDB Molecule:thit;
PDBTitle: crystal structure at 2.0 a of the s-component for thiamin from an ecf-2 type abc transporter
Phyre2
| 17 |
|
PDB 2jp3 chain A
Region: 324 - 367
Aligned: 44
Modelled: 44
Confidence: 6.1%
Identity: 14%
PDB header:transcription
Chain: A: PDB Molecule:fxyd domain-containing ion transport regulator 4;
PDBTitle: solution structure of the human fxyd4 (chif) protein in sds2 micelles
Phyre2
| 18 |
|
PDB 2kb1 chain A
Region: 319 - 354
Aligned: 36
Modelled: 36
Confidence: 5.7%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:wsk3;
PDBTitle: nmr studies of a channel protein without membrane:2 structure and dynamics of water-solubilized kcsa
Phyre2
| 19 |
|
PDB 1iwg chain A domain 8
Region: 277 - 361
Aligned: 85
Modelled: 85
Confidence: 5.7%
Identity: 8%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 20 |
|
PDB 2ww9 chain B
Region: 311 - 337
Aligned: 27
Modelled: 27
Confidence: 5.6%
Identity: 37%
PDB header:ribosome
Chain: B: PDB Molecule:protein transport protein sss1;
PDBTitle: cryo-em structure of the active yeast ssh1 complex bound to the2 yeast 80s ribosome
Phyre2