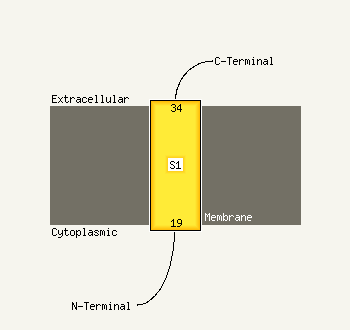
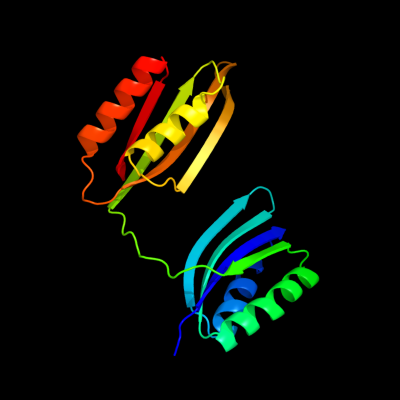
| 1 | c1u8sB_
|
|
 |
100.0 |
44 |
PDB header:transcription
Chain: B: PDB Molecule:glycine cleavage system transcriptional
PDBTitle: crystal structure of putative glycine cleavage system2 transcriptional repressor
|
| 2 | c2nyiB_
|
|
 |
100.0 |
24 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:unknown protein;
PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
|
| 3 | c3p96A_
|
|
 |
99.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphoserine phosphatase serb;
PDBTitle: crystal structure of phosphoserine phosphatase serb from mycobacterium2 avium, native form
|
| 4 | d1u8sa1
|
|
 |
99.8 |
55 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 5 | c3nrbD_
|
|
 |
99.7 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (puru,2 pp_1943) from pseudomonas putida kt2440 at 2.05 a resolution
|
| 6 | d1u8sa2
|
|
 |
99.7 |
34 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 7 | d1zpva1
|
|
 |
99.7 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:SP0238-like |
| 8 | c3obiC_
|
|
 |
99.6 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (np_949368)2 from rhodopseudomonas palustris cga009 at 1.95 a resolution
|
| 9 | c3n0vD_
|
|
 |
99.6 |
12 |
PDB header:hydrolase
Chain: D: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pp_0327)2 from pseudomonas putida kt2440 at 2.25 a resolution
|
| 10 | c3louB_
|
|
 |
99.6 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of formyltetrahydrofolate deformylase (yp_105254.1)2 from burkholderia mallei atcc 23344 at 1.90 a resolution
|
| 11 | c3o1lB_
|
|
 |
99.6 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:formyltetrahydrofolate deformylase;
PDBTitle: crystal structure of a formyltetrahydrofolate deformylase (pspto_4314)2 from pseudomonas syringae pv. tomato str. dc3000 at 2.20 a resolution
|
| 12 | c3l76B_
|
|
 |
99.0 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of aspartate kinase from synechocystis
|
| 13 | c2f06B_
|
|
 |
99.0 |
12 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:conserved hypothetical protein;
PDBTitle: crystal structure of protein bt0572 from bacteroides thetaiotaomicron
|
| 14 | c3ibwA_
|
|
 |
98.4 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:gtp pyrophosphokinase;
PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
|
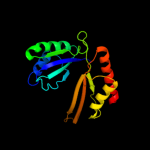
| 15 | c2dtjA_
|
|
 |
98.3 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of regulatory subunit of aspartate kinase2 from corynebacterium glutamicum
|
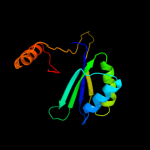
| 16 | c2zhoB_
|
|
 |
98.2 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of the regulatory subunit of aspartate2 kinase from thermus thermophilus (ligand free form)
|
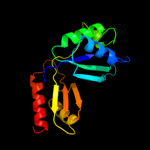
| 17 | c2re1A_
|
|
 |
98.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase, alpha and beta subunits;
PDBTitle: crystal structure of aspartokinase alpha and beta subunits
|
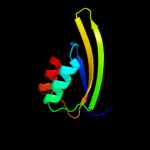
| 18 | d1ygya3
|
|
 |
97.9 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 19 | d1sc6a3
|
|
 |
97.8 |
10 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phosphoglycerate dehydrogenase, regulatory (C-terminal) domain |
| 20 | d2fgca2
|
|
 |
97.8 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 21 | c3c1nA_ |
|
not modelled |
97.8 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:probable aspartokinase;
PDBTitle: crystal structure of allosteric inhibition threonine-sensitive2 aspartokinase from methanococcus jannaschii with l-threonine
|
| 22 | d2f06a2 |
|
not modelled |
97.7 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 23 | c2fgcA_ |
|
not modelled |
97.7 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase, small subunit;
PDBTitle: crystal structure of acetolactate synthase- small subunit from2 thermotoga maritima
|
| 24 | d2pc6a2 |
|
not modelled |
97.7 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 25 | d2f1fa1 |
|
not modelled |
97.7 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:IlvH-like |
| 26 | c2pc6C_ |
|
not modelled |
97.6 |
15 |
PDB header:lyase
Chain: C: PDB Molecule:probable acetolactate synthase isozyme iii (small subunit);
PDBTitle: crystal structure of putative acetolactate synthase- small subunit2 from nitrosomonas europaea
|
| 27 | c3mwbA_ |
|
not modelled |
97.5 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of prephenate dehydratase in complex with l-phe2 from arthrobacter aurescens to 2.0a
|
| 28 | c3ab4K_ |
|
not modelled |
97.3 |
15 |
PDB header:transferase
Chain: K: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of feedback inhibition resistant mutant of aspartate2 kinase from corynebacterium glutamicum in complex with lysine and3 threonine
|
| 29 | c2f1fA_ |
|
not modelled |
97.3 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:acetolactate synthase isozyme iii small subunit;
PDBTitle: crystal structure of the regulatory subunit of2 acetohydroxyacid synthase isozyme iii from e. coli
|
| 30 | c2cdqB_ |
|
not modelled |
97.2 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:aspartokinase;
PDBTitle: crystal structure of arabidopsis thaliana aspartate kinase2 complexed with lysine and s-adenosylmethionine
|
| 31 | d2f06a1 |
|
not modelled |
97.2 |
16 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:BT0572-like |
| 32 | d2qmwa2 |
|
not modelled |
97.1 |
17 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 33 | c2qmxB_ |
|
not modelled |
97.0 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of l-phe inhibited prephenate dehydratase from2 chlorobium tepidum tls
|
| 34 | c1ygyA_ |
|
not modelled |
97.0 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
|
| 35 | c2qmwA_ |
|
not modelled |
96.9 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:prephenate dehydratase;
PDBTitle: the crystal structure of the prephenate dehydratase (pdt) from2 staphylococcus aureus subsp. aureus mu50
|
| 36 | c3luyA_ |
|
not modelled |
96.7 |
13 |
PDB header:isomerase
Chain: A: PDB Molecule:probable chorismate mutase;
PDBTitle: putative chorismate mutase from bifidobacterium adolescentis
|
| 37 | d1phza1 |
|
not modelled |
96.6 |
8 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Phenylalanine metabolism regulatory domain |
| 38 | c3mtjA_ |
|
not modelled |
96.1 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: the crystal structure of a homoserine dehydrogenase from thiobacillus2 denitrificans to 2.15a
|
| 39 | c3mahA_ |
|
not modelled |
96.0 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:aspartokinase;
PDBTitle: a putative c-terminal regulatory domain of aspartate kinase from2 porphyromonas gingivalis w83.
|
| 40 | d2hmfa3 |
|
not modelled |
95.6 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 41 | c1y7pB_ |
|
not modelled |
94.8 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein af1403;
PDBTitle: 1.9 a crystal structure of a protein of unknown function2 af1403 from archaeoglobus fulgidus, probable metabolic3 regulator
|
| 42 | c3k5pA_ |
|
not modelled |
94.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: crystal structure of amino acid-binding act: d-isomer specific 2-2 hydroxyacid dehydrogenase catalytic domain from brucella melitensis
|
| 43 | d2hmfa2 |
|
not modelled |
94.1 |
24 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 44 | c1ybaC_ |
|
not modelled |
93.6 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:d-3-phosphoglycerate dehydrogenase;
PDBTitle: the active form of phosphoglycerate dehydrogenase
|
| 45 | c2j0wA_ |
|
not modelled |
92.7 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:lysine-sensitive aspartokinase 3;
PDBTitle: crystal structure of e. coli aspartokinase iii in complex2 with aspartate and adp (r-state)
|
| 46 | d2cdqa2 |
|
not modelled |
92.3 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 47 | d2cdqa3 |
|
not modelled |
86.4 |
15 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 48 | d2j0wa2 |
|
not modelled |
86.4 |
11 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 49 | c1tdjA_ |
|
not modelled |
79.7 |
9 |
PDB header:allostery
Chain: A: PDB Molecule:biosynthetic threonine deaminase;
PDBTitle: threonine deaminase (biosynthetic) from e. coli
|
| 50 | c2phmA_ |
|
not modelled |
73.2 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (phenylalanine-4-hydroxylase);
PDBTitle: structure of phenylalanine hydroxylase dephosphorylated
|
| 51 | d1vgya2 |
|
not modelled |
60.0 |
14 |
Fold:Ferredoxin-like
Superfamily:Bacterial exopeptidase dimerisation domain
Family:Bacterial exopeptidase dimerisation domain |
| 52 | c2jheB_ |
|
not modelled |
59.8 |
12 |
PDB header:transcription
Chain: B: PDB Molecule:transcription regulator tyrr;
PDBTitle: n-terminal domain of tyrr transcription factor (residues 1 -2 190)
|
| 53 | d2j0wa3 |
|
not modelled |
56.4 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Aspartokinase allosteric domain-like |
| 54 | d1zhva2 |
|
not modelled |
52.3 |
7 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Atu0741-like |
| 55 | d1rwua_ |
|
not modelled |
51.7 |
14 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:YbeD-like |
| 56 | c1rwuA_ |
|
not modelled |
51.7 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical upf0250 protein ybed;
PDBTitle: solution structure of conserved protein ybed from e. coli
|
| 57 | d3ceda1 |
|
not modelled |
49.4 |
9 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:NIL domain-like |
| 58 | c1zhvA_ |
|
not modelled |
46.9 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein atu0741;
PDBTitle: x-ray crystal structure protein atu0741 from agobacterium tumefaciens.2 northeast structural genomics consortium target atr8.
|
| 59 | d1geea_ |
|
not modelled |
43.6 |
11 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 60 | d1zvpa2 |
|
not modelled |
38.6 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:VC0802-like |
| 61 | d1gtda_ |
|
not modelled |
36.3 |
8 |
Fold:PurS-like
Superfamily:PurS-like
Family:PurS subunit of FGAM synthetase |
| 62 | c2qc8J_ |
|
not modelled |
34.6 |
19 |
PDB header:ligase
Chain: J: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of human glutamine synthetase in complex with adp2 and methionine sulfoximine phosphate
|
| 63 | c2yvqA_ |
|
not modelled |
33.6 |
11 |
PDB header:ligase
Chain: A: PDB Molecule:carbamoyl-phosphate synthase;
PDBTitle: crystal structure of mgs domain of carbamoyl-phosphate2 synthetase from homo sapiens
|
| 64 | d1v5va2 |
|
not modelled |
32.0 |
12 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
| 65 | c1zvpB_ |
|
not modelled |
31.8 |
9 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical protein vc0802;
PDBTitle: crystal structure of a protein of unknown function vc0802 from vibrio2 cholerae, possible transport protein
|
| 66 | c2uygF_ |
|
not modelled |
30.8 |
19 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 67 | d3bypa1 |
|
not modelled |
29.9 |
15 |
Fold:Alpha-lytic protease prodomain-like
Superfamily:Cation efflux protein cytoplasmic domain-like
Family:Cation efflux protein cytoplasmic domain-like |
| 68 | d1gqoa_ |
|
not modelled |
28.7 |
16 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 69 | c3n8kG_ |
|
not modelled |
25.6 |
6 |
PDB header:lyase
Chain: G: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: type ii dehydroquinase from mycobacterium tuberculosis complexed with2 citrazinic acid
|
| 70 | d1wosa2 |
|
not modelled |
25.1 |
9 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
| 71 | d1uqra_ |
|
not modelled |
23.6 |
16 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 72 | c3lwzC_ |
|
not modelled |
23.1 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.65 angstrom resolution crystal structure of type ii 3-2 dehydroquinate dehydratase (aroq) from yersinia pestis
|
| 73 | d1u5ta1 |
|
not modelled |
22.7 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Vacuolar sorting protein domain |
| 74 | c1x31A_ |
|
not modelled |
22.5 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sarcosine oxidase alpha subunit;
PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
|
| 75 | d1lfpa_ |
|
not modelled |
22.4 |
14 |
Fold:YebC-like
Superfamily:YebC-like
Family:YebC-like |
| 76 | c1worA_ |
|
not modelled |
21.9 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:aminomethyltransferase;
PDBTitle: crystal structure of t-protein of the glycine cleavage2 system
|
| 77 | d1fc4a_ |
|
not modelled |
20.9 |
13 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:GABA-aminotransferase-like |
| 78 | c2k4mA_ |
|
not modelled |
19.9 |
11 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0146 protein mth_1000;
PDBTitle: solution nmr structure of m. thermoautotrophicum protein2 mth_1000, northeast structural genomics consortium target3 tr8
|
| 79 | d1zcza1 |
|
not modelled |
19.4 |
17 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Inosicase |
| 80 | d1xq1a_ |
|
not modelled |
17.6 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 81 | d1gtza_ |
|
not modelled |
17.3 |
13 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 82 | c3u80A_ |
|
not modelled |
16.7 |
4 |
PDB header:unknown function
Chain: A: PDB Molecule:3-dehydroquinate dehydratase, type ii;
PDBTitle: 1.60 angstrom resolution crystal structure of a 3-dehydroquinate2 dehydratase-like protein from bifidobacterium longum
|
| 83 | c2oz5A_ |
|
not modelled |
14.7 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:phosphotyrosine protein phosphatase ptpb;
PDBTitle: crystal structure of mycobacterium tuberculosis protein tyrosine2 phosphatase ptpb in complex with the specific inhibitor omts
|
| 84 | d1ulua_ |
|
not modelled |
14.5 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 85 | d1pj5a4 |
|
not modelled |
14.5 |
13 |
Fold:Folate-binding domain
Superfamily:Folate-binding domain
Family:Aminomethyltransferase folate-binding domain |
| 86 | c2ptgA_ |
|
not modelled |
14.5 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: crystal structure of eimeria tenella enoyl reductase
|
| 87 | d1h05a_ |
|
not modelled |
13.9 |
7 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 88 | d1nxia_ |
|
not modelled |
13.1 |
11 |
Fold:Ferredoxin-like
Superfamily:Hypothetical protein VC0424
Family:Hypothetical protein VC0424 |
| 89 | c2d1uA_ |
|
not modelled |
12.4 |
7 |
PDB header:metal transport
Chain: A: PDB Molecule:iron(iii) dicitrate transport protein feca;
PDBTitle: solution strcuture of the periplasmic signaling domain of2 feca from escherichia coli
|
| 90 | d1tdja2 |
|
not modelled |
11.6 |
12 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Allosteric threonine deaminase C-terminal domain |
| 91 | d2joqa1 |
|
not modelled |
11.5 |
3 |
Fold:Ferredoxin-like
Superfamily:YbeD/HP0495-like
Family:HP0495-like |
| 92 | c2zw2B_ |
|
not modelled |
11.2 |
16 |
PDB header:ligase
Chain: B: PDB Molecule:putative uncharacterized protein sts178;
PDBTitle: crystal structure of formylglycinamide ribonucleotide amidotransferase2 iii from sulfolobus tokodaii (stpurs)
|
| 93 | c3jrnA_ |
|
not modelled |
11.1 |
9 |
PDB header:plant protein
Chain: A: PDB Molecule:at1g72930 protein;
PDBTitle: crystal structure of tir domain from arabidopsis thaliana
|
| 94 | c3oziB_ |
|
not modelled |
11.0 |
9 |
PDB header:plant protein
Chain: B: PDB Molecule:l6tr;
PDBTitle: crystal structure of the tir domain from the flax disease resistance2 protein l6
|
| 95 | d1d7oa_ |
|
not modelled |
10.8 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
| 96 | c2zmeA_ |
|
not modelled |
10.7 |
12 |
PDB header:protein transport
Chain: A: PDB Molecule:vacuolar-sorting protein snf8;
PDBTitle: integrated structural and functional model of the human escrt-ii2 complex
|
| 97 | d1a9xa2 |
|
not modelled |
10.6 |
11 |
Fold:Methylglyoxal synthase-like
Superfamily:Methylglyoxal synthase-like
Family:Carbamoyl phosphate synthetase, large subunit allosteric, C-terminal domain |
| 98 | d1ywfa1 |
|
not modelled |
10.2 |
33 |
Fold:(Phosphotyrosine protein) phosphatases II
Superfamily:(Phosphotyrosine protein) phosphatases II
Family:Mycobacterial PtpB-like |
| 99 | c2w8wA_ |
|
not modelled |
9.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:serine palmitoyltransferase;
PDBTitle: n100y spt with plp-ser
|