| 1 |
|
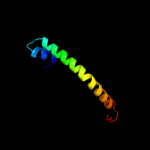
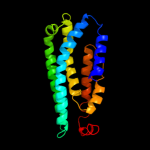
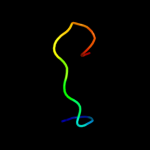
PDB 3qnq chain D
Region: 207 - 261
Aligned: 55
Modelled: 55
Confidence: 38.0%
Identity: 18%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 2 |
|
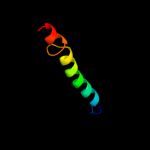
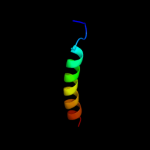
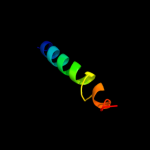
PDB 2l34 chain B
Region: 31 - 65
Aligned: 33
Modelled: 34
Confidence: 32.0%
Identity: 18%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 3 |
|
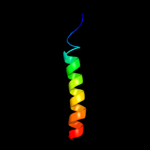
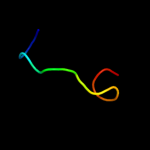
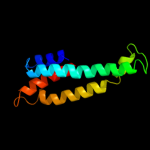
PDB 2kdx chain A
Region: 1 - 11
Aligned: 11
Modelled: 11
Confidence: 31.0%
Identity: 36%
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
Phyre2
| 4 |
|
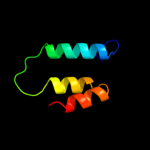
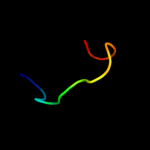
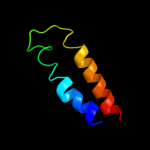
PDB 2l35 chain B
Region: 31 - 57
Aligned: 27
Modelled: 27
Confidence: 29.4%
Identity: 15%
PDB header:protein binding
Chain: B: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 5 |
|
PDB 1cii chain A
Region: 70 - 121
Aligned: 51
Modelled: 52
Confidence: 23.6%
Identity: 18%
PDB header:transmembrane protein
Chain: A: PDB Molecule:colicin ia;
PDBTitle: colicin ia
Phyre2
| 6 |
|
PDB 3mku chain A
Region: 74 - 255
Aligned: 182
Modelled: 182
Confidence: 22.8%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
Phyre2
| 7 |
|
PDB 2l34 chain A
Region: 31 - 57
Aligned: 27
Modelled: 27
Confidence: 20.5%
Identity: 15%
PDB header:protein binding
Chain: A: PDB Molecule:tyro protein tyrosine kinase-binding protein;
PDBTitle: structure of the dap12 transmembrane homodimer
Phyre2
| 8 |
|
PDB 2eel chain A
Region: 252 - 265
Aligned: 14
Modelled: 14
Confidence: 19.3%
Identity: 7%
PDB header:apoptosis
Chain: A: PDB Molecule:cell death activator cide-a;
PDBTitle: solution structure of the cide-n domain of human cell death2 activator cide-a
Phyre2
| 9 |
|
PDB 1ibx chain B
Region: 252 - 264
Aligned: 13
Modelled: 13
Confidence: 15.4%
Identity: 15%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: CAD domain
Phyre2
| 10 |
|
PDB 1ibx chain B
Region: 252 - 264
Aligned: 13
Modelled: 13
Confidence: 15.4%
Identity: 15%
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:chimera of igg binding protein g and dna
PDBTitle: nmr structure of dff40 and dff45 n-terminal domain complex
Phyre2
| 11 |
|
PDB 1d4b chain A
Region: 252 - 264
Aligned: 13
Modelled: 13
Confidence: 14.8%
Identity: 15%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: CAD domain
Phyre2
| 12 |
|
PDB 2l35 chain A
Region: 37 - 66
Aligned: 28
Modelled: 30
Confidence: 14.0%
Identity: 21%
PDB header:protein binding
Chain: A: PDB Molecule:dap12-nkg2c_tm;
PDBTitle: structure of the dap12-nkg2c transmembrane heterotrimer
Phyre2
| 13 |
|
PDB 1f2r chain I
Region: 252 - 265
Aligned: 14
Modelled: 14
Confidence: 12.6%
Identity: 14%
Fold: beta-Grasp (ubiquitin-like)
Superfamily: CAD & PB1 domains
Family: CAD domain
Phyre2
| 14 |
|
PDB 1iwg chain A domain 8
Region: 79 - 185
Aligned: 107
Modelled: 107
Confidence: 10.5%
Identity: 7%
Fold: Multidrug efflux transporter AcrB transmembrane domain
Superfamily: Multidrug efflux transporter AcrB transmembrane domain
Family: Multidrug efflux transporter AcrB transmembrane domain
Phyre2
| 15 |
|
PDB 1fft chain C
Region: 109 - 154
Aligned: 46
Modelled: 46
Confidence: 9.1%
Identity: 15%
Fold: Cytochrome c oxidase subunit III-like
Superfamily: Cytochrome c oxidase subunit III-like
Family: Cytochrome c oxidase subunit III-like
Phyre2
| 16 |
|
PDB 1v54 chain I
Region: 215 - 264
Aligned: 50
Modelled: 50
Confidence: 7.2%
Identity: 16%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIc
Family: Mitochondrial cytochrome c oxidase subunit VIc
Phyre2
| 17 |
|
PDB 1eys chain H domain 2
Region: 219 - 249
Aligned: 31
Modelled: 31
Confidence: 7.1%
Identity: 13%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction centre subunit H, transmembrane region
Family: Photosystem II reaction centre subunit H, transmembrane region
Phyre2
| 18 |
|
PDB 2jo1 chain A
Region: 225 - 267
Aligned: 43
Modelled: 43
Confidence: 6.8%
Identity: 12%
PDB header:hydrolase regulator
Chain: A: PDB Molecule:phospholemman;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
Phyre2
| 19 |
|
PDB 2ko6 chain A
Region: 236 - 263
Aligned: 28
Modelled: 28
Confidence: 6.5%
Identity: 18%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein yihd;
PDBTitle: solution structure of protein sf3929 from shigella flexneri2 2a. northeast structural genomics consortium target3 sfr81/ontario center for structural proteomics target4 sf3929
Phyre2
| 20 |
|
PDB 1r7m chain A domain 1
Region: 33 - 46
Aligned: 14
Modelled: 14
Confidence: 5.7%
Identity: 36%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 21 |
|