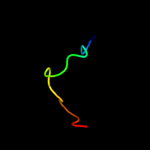
1 d2enga_
44.9
53
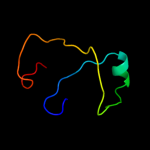
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: Eng V-like2 d1l8fa_
42.4
50
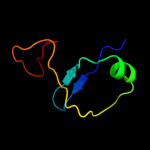
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: Eng V-like3 d1miua3
37.0
23
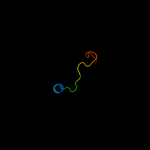
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB4 d1iyjb3
36.2
22
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Single strand DNA-binding domain, SSB5 d1j6xa_
34.2
35
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS6 d1vjea_
29.7
42
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS7 d1i5pa1
25.6
38
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: delta-Endotoxin, C-terminal domain8 d1j98a_
24.0
38
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS9 d1j6wa_
22.6
46
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: Autoinducer-2 production protein LuxS10 c2qqpD_
22.4
48
PDB header: virusChain: D: PDB Molecule: small capsid protein;PDBTitle: crystal structure of authentic providence virus
11 d1wy5a2
21.3
24
Fold: MesJ substrate recognition domain-likeSuperfamily: MesJ substrate recognition domain-likeFamily: MesJ substrate recognition domain-like12 d1cfra_
16.8
23
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Cfr10I/Bse634I13 c3na2C_
14.0
30
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function from mine drainage2 metagenome leptospirillum rubarum
14 d1tbfa_
13.4
27
Fold: HD-domain/PDEase-likeSuperfamily: HD-domain/PDEase-likeFamily: PDEase15 c2fvuA_
13.2
41
PDB header: transcriptionChain: A: PDB Molecule: regulatory protein sir3;PDBTitle: structure of the yeast sir3 bah domain
16 c1xozA_
13.0
27
PDB header: hydrolaseChain: A: PDB Molecule: cgmp-specific 3',5'-cyclic phosphodiesterase;PDBTitle: catalytic domain of human phosphodiesterase 5a in complex2 with tadalafil
17 d1qmja_
12.4
27
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)18 c2cosA_
11.7
88
PDB header: transferaseChain: A: PDB Molecule: serine/threonine protein kinase lats2;PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
19 c2vy8A_
11.1
40
PDB header: transcriptionChain: A: PDB Molecule: polymerase basic protein 2;PDBTitle: the 627-domain from influenza a virus polymerase pb22 subunit with glu-627
20 d1z0kb1
9.9
50
Fold: Long alpha-hairpinSuperfamily: Rabenosyn-5 Rab-binding domain-likeFamily: Rabenosyn-5 Rab-binding domain-like21 c1mjeA_
not modelled
9.8
23
PDB header: gene regulation/antitumor protein/dnaChain: A: PDB Molecule: breast cancer 2;PDBTitle: structure of a brca2-dss1-ssdna complex
22 d1ddba_
not modelled
9.6
30
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death23 d1yzma1
not modelled
9.5
50
Fold: Long alpha-hairpinSuperfamily: Rabenosyn-5 Rab-binding domain-likeFamily: Rabenosyn-5 Rab-binding domain-like24 c3cw4A_
not modelled
9.2
40
PDB header: transferaseChain: A: PDB Molecule: polymerase basic protein 2;PDBTitle: large c-terminal domain of influenza a virus rna-dependent polymerase2 pb2
25 c1x50A_
not modelled
9.0
16
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-4;PDBTitle: solution structure of the c-terminal gal-bind lectin domain2 from human galectin-4
26 c2q2kA_
not modelled
9.0
58
PDB header: dna binding protein/dnaChain: A: PDB Molecule: hypothetical protein;PDBTitle: structure of nucleic-acid binding protein
27 c3c8lB_
not modelled
8.8
29
PDB header: unknown functionChain: B: PDB Molecule: ftsz-like protein of unknown function;PDBTitle: crystal structure of a ftsz-like protein of unknown function2 (npun_r1471) from nostoc punctiforme pcc 73102 at 1.22 a resolution
28 c2wg6L_
not modelled
8.4
24
PDB header: transcription,hydrolaseChain: L: PDB Molecule: general control protein gcn4,PDBTitle: proteasome-activating nucleotidase (pan) n-domain (57-134)2 from archaeoglobus fulgidus fused to gcn4, p61a mutant
29 d1hlca_
not modelled
8.4
21
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Galectin (animal S-lectin)30 c3duiB_
not modelled
7.5
20
PDB header: sugar binding proteinChain: B: PDB Molecule: beta-galactoside-binding lectin;PDBTitle: crystal structure of the oxidized cg-1b: an adhesion/growth-2 regulatory lectin from chicken
31 d2cosa1
not modelled
7.4
88
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain32 d1ok7a2
not modelled
7.2
17
Fold: DNA clampSuperfamily: DNA clampFamily: DNA polymerase III, beta subunit33 c3h6nA_
not modelled
6.7
21
PDB header: signaling proteinChain: A: PDB Molecule: plexin-d1;PDBTitle: crystal structure of the ubiquitin-like domain of plexin d1
34 c3bjcA_
not modelled
6.5
23
PDB header: hydrolaseChain: A: PDB Molecule: cgmp-specific 3',5'-cyclic phosphodiesterase;PDBTitle: crystal structure of the pde5a catalytic domain in complex2 with a novel inhibitor
35 c1iyjB_
not modelled
6.5
22
PDB header: gene regulation/antitumor proteinChain: B: PDB Molecule: breast cancer susceptibility;PDBTitle: structure of a brca2-dss1 complex
36 c3najA_
not modelled
6.3
18
PDB header: sugar binding proteinChain: A: PDB Molecule: galectin-8;PDBTitle: crystal structure of a protease-resistant mutant form of human2 galectin-8
37 c3zwfA_
not modelled
6.3
36
PDB header: hydrolaseChain: A: PDB Molecule: zinc phosphodiesterase elac protein 1;PDBTitle: crystal structure of human trnase z, short form (elac1).
38 c3bg4D_
not modelled
6.2
30
PDB header: hydrolase/hydrolase inhibitorChain: D: PDB Molecule: guamerin;PDBTitle: the crystal structure of guamerin in complex with2 chymotrypsin and the development of an elastase-specific3 inhibitor
39 d1f3va_
not modelled
6.0
44
Fold: Ferredoxin-likeSuperfamily: TRADD, N-terminal domainFamily: TRADD, N-terminal domain40 c2l6pA_
not modelled
6.0
19
PDB header: structure genomics, unknown functionChain: A: PDB Molecule: phac1, phac2 and phad genes;PDBTitle: nmr solution structure of the protein np_253742.1
41 c4a0kB_
not modelled
5.9
83
PDB header: ligase/dna-binding protein/dnaChain: B: PDB Molecule: e3 ubiquitin-protein ligase rbx1;PDBTitle: structure of ddb1-ddb2-cul4a-rbx1 bound to a 12 bp abasic2 site containing dna-duplex
42 c3phfX_
not modelled
5.8
23
PDB header: viral proteinChain: X: PDB Molecule: envelope glycoprotein l;PDBTitle: crystal structure of the epstein-barr virus gh and gl complex
43 c1gl9B_
not modelled
5.7
22
PDB header: topoisomeraseChain: B: PDB Molecule: reverse gyrase;PDBTitle: archaeoglobus fulgidus reverse gyrase complexed with adpnp
44 c2jphA_
not modelled
5.7
21
PDB header: signaling protein, protein bindingChain: A: PDB Molecule: plexin-b1;PDBTitle: nmr solution structure of the rho gtpase binding domain of2 human plexin-b1
45 c1xtcC_
not modelled
5.6
47
PDB header: toxinChain: C: PDB Molecule: cholera toxin;PDBTitle: cholera toxin
46 c3h43F_
not modelled
5.6
19
PDB header: hydrolaseChain: F: PDB Molecule: proteasome-activating nucleotidase;PDBTitle: n-terminal domain of the proteasome-activating nucleotidase2 of methanocaldococcus jannaschii
47 c2ounA_
not modelled
5.2
24
PDB header: hydrolaseChain: A: PDB Molecule: camp and camp-inhibited cgmp 3',5'-cyclicPDBTitle: crystal structure of pde10a2 in complex with amp