1 c1yf5L_
99.9
16
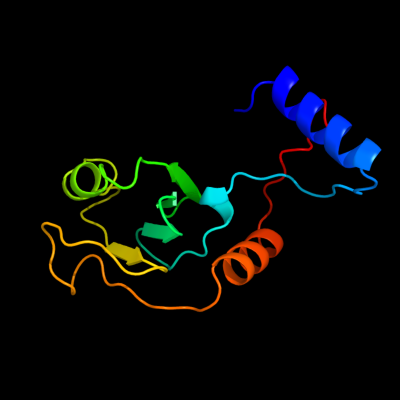
PDB header: transport proteinChain: L: PDB Molecule: general secretion pathway protein l;PDBTitle: cyto-epsl: the cytoplasmic domain of epsl, an inner membrane component2 of the type ii secretion system of vibrio cholerae
2 c2w7vB_
99.8
29
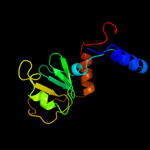
PDB header: transport proteinChain: B: PDB Molecule: general secretion pathway protein l;PDBTitle: periplasmic domain of epsl from vibrio parahaemolyticus
3 d2bh1a1
99.6
15
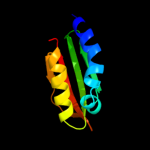
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Cyto-EpsL domain4 d1uv7a_
78.8
11
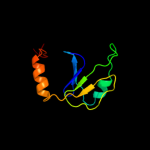
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: General secretion pathway protein M, EpsMFamily: General secretion pathway protein M, EpsM5 c1uv7A_
78.8
11
PDB header: transportChain: A: PDB Molecule: general secretion pathway protein m;PDBTitle: periplasmic domain of epsm from vibrio cholerae
6 c2l3mA_
33.8
20
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
7 d1p6ta2
29.6
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain8 d2aw0a_
27.1
10
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain9 d1cpza_
21.5
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain10 d1s6ua_
20.9
4
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain11 d1sb6a_
20.5
13
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain12 c2kt2A_
17.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
13 c2jpiA_
17.3
13
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical protein;PDBTitle: chemical shift assignments of pa4090 from pseudomonas2 aeruginosa
14 d1p6ta1
17.1
10
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain15 d2qifa1
16.0
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain16 d1f44a1
14.5
19
Fold: SAM domain-likeSuperfamily: lambda integrase-like, N-terminal domainFamily: lambda integrase-like, N-terminal domain17 c2k2pA_
14.4
6
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
18 c3dxsX_
13.7
4
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
19 d1mwza_
13.5
10
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain20 d1j5ya2
13.2
8
Fold: HPr-likeSuperfamily: Putative transcriptional regulator TM1602, C-terminal domainFamily: Putative transcriptional regulator TM1602, C-terminal domain21 c1yg0A_
not modelled
12.9
14
PDB header: metal transportChain: A: PDB Molecule: cop associated protein;PDBTitle: solution structure of apo-copp from helicobacter pylori
22 c2x29A_
not modelled
11.9
22
PDB header: cell adhesionChain: A: PDB Molecule: tumor necrosis factor ligand superfamily memberPDBTitle: crystal structure of human4-1bb ligand ectodomain
23 d1kvja_
not modelled
11.5
4
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain24 c3hd7A_
not modelled
11.1
12
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
25 d2ggpb1
not modelled
10.7
3
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain26 c2gcfA_
not modelled
9.8
5
PDB header: hydrolaseChain: A: PDB Molecule: cation-transporting atpase pacs;PDBTitle: solution structure of the n-terminal domain of the coppper(i) atpase2 pacs in its apo form
27 c3ibwA_
not modelled
9.6
17
PDB header: transferaseChain: A: PDB Molecule: gtp pyrophosphokinase;PDBTitle: crystal structure of the act domain from gtp2 pyrophosphokinase of chlorobium tepidum. northeast3 structural genomics consortium target ctr148a
28 c2rrlA_
not modelled
9.4
16
PDB header: protein transportChain: A: PDB Molecule: flagellar hook-length control protein;PDBTitle: solution structure of the c-terminal domain of the flik
29 d1je3a_
not modelled
9.2
15
Fold: IF3-likeSuperfamily: SirA-likeFamily: SirA-like30 c2kyzA_
not modelled
9.1
15
PDB header: metal binding proteinChain: A: PDB Molecule: heavy metal binding protein;PDBTitle: nmr structure of heavy metal binding protein tm0320 from thermotoga2 maritima
31 d2vzsa2
not modelled
9.0
15
Fold: Immunoglobulin-like beta-sandwichSuperfamily: beta-Galactosidase/glucuronidase domainFamily: beta-Galactosidase/glucuronidase domain32 c2ofhX_
not modelled
9.0
11
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
33 c2yciX_
not modelled
8.8
16
PDB header: transferaseChain: X: PDB Molecule: 5-methyltetrahydrofolate corrinoid/iron sulfur proteinPDBTitle: methyltransferase native
34 d1osda_
not modelled
8.3
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain35 c2l3wA_
not modelled
7.7
7
PDB header: photosynthesisChain: A: PDB Molecule: phycobilisome rod linker polypeptide;PDBTitle: solution nmr structure of the pbs linker domain of phycobilisome rod2 linker polypeptide from synechococcus elongatus, northeast structural3 genomics consortium target snr168a
36 c3h0dB_
not modelled
7.5
25
PDB header: transcription/dnaChain: B: PDB Molecule: ctsr;PDBTitle: crystal structure of ctsr in complex with a 26bp dna duplex
37 d1afia_
not modelled
7.3
15
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain38 c3k13A_
not modelled
7.2
9
PDB header: transferaseChain: A: PDB Molecule: 5-methyltetrahydrofolate-homocysteine methyltransferase;PDBTitle: structure of the pterin-binding domain metr of 5-2 methyltetrahydrofolate-homocysteine methyltransferase from3 bacteroides thetaiotaomicron
39 c1yjrA_
not modelled
7.1
3
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
40 c3fryB_
not modelled
7.1
14
PDB header: hydrolaseChain: B: PDB Molecule: probable copper-exporting p-type atpase a;PDBTitle: crystal structure of the copa c-terminal metal binding domain
41 c3nklA_
not modelled
6.9
14
PDB header: oxidoreductase/lyaseChain: A: PDB Molecule: udp-d-quinovosamine 4-dehydrogenase;PDBTitle: crystal structure of udp-d-quinovosamine 4-dehydrogenase from vibrio2 fischeri
42 c2rjzA_
not modelled
6.9
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: pilo protein;PDBTitle: crystal structure of the type 4 fimbrial biogenesis protein pilo from2 pseudomonas aeruginosa
43 c2ldiA_
not modelled
6.5
11
PDB header: hydrolaseChain: A: PDB Molecule: zinc-transporting atpase;PDBTitle: nmr solution structure of ziaan sub mutant
44 c2ropA_
not modelled
6.5
14
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 2;PDBTitle: solution structure of domains 3 and 4 of human atp7b
45 c2ychA_
not modelled
6.5
10
PDB header: cell cycleChain: A: PDB Molecule: competence protein pilm;PDBTitle: pilm-piln type iv pilus biogenesis complex
46 d1q8la_
not modelled
6.2
5
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain47 c1y3kA_
not modelled
5.8
8
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the fifth domain of2 menkes protein
48 c2k42B_
not modelled
5.4
30
PDB header: signaling proteinChain: B: PDB Molecule: espfu;PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
49 d1t1ea2
not modelled
5.1
10
Fold: Ferredoxin-likeSuperfamily: Protease propeptides/inhibitorsFamily: Subtilase propeptides/inhibitors