1 c3rkoL_
100.0
32
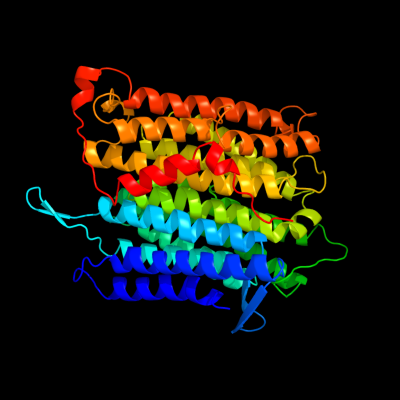
PDB header: oxidoreductaseChain: L: PDB Molecule: nadh-quinone oxidoreductase subunit l;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
2 c3rkoM_
100.0
19
PDB header: oxidoreductaseChain: M: PDB Molecule: nadh-quinone oxidoreductase subunit m;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
3 c3rkoN_
100.0
19
PDB header: oxidoreductaseChain: N: PDB Molecule: nadh-quinone oxidoreductase subunit n;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
4 c3rkoK_
61.3
20
PDB header: oxidoreductaseChain: K: PDB Molecule: nadh-quinone oxidoreductase subunit k;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
5 c2l3iA_
33.3
50
PDB header: antimicrobial proteinChain: A: PDB Molecule: aoxki4a, antimicrobial peptide in spider venom;PDBTitle: oxki4a, spider derived antimicrobial peptide
6 d1a6qa1
24.6
26
Fold: Another 3-helical bundleSuperfamily: Protein serine/threonine phosphatase 2C, C-terminal domainFamily: Protein serine/threonine phosphatase 2C, C-terminal domain7 c3r24A_
14.9
22
PDB header: transferase, viral proteinChain: A: PDB Molecule: 2'-o-methyl transferase;PDBTitle: crystal structure of nsp10/nsp16 complex of sars coronavirus" if2 possible
8 d1s1qa_
14.6
31
Fold: UBC-likeSuperfamily: UBC-likeFamily: UEV domain9 d1n9wa2
14.5
63
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain10 c1n9wA_
14.5
63
PDB header: biosynthetic proteinChain: A: PDB Molecule: aspartyl-trna synthetase 2;PDBTitle: crystal structure of the non-discriminating and archaeal-2 type aspartyl-trna synthetase from thermus thermophilus
11 c2kseA_
14.0
19
PDB header: transferaseChain: A: PDB Molecule: sensor protein qsec;PDBTitle: backbone structure of the membrane domain of e. coli2 histidine kinase receptor qsec, center for structures of3 membrane proteins (csmp) target 4311c
12 c2k21A_
13.9
28
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
13 c3bjuB_
13.8
50
PDB header: ligaseChain: B: PDB Molecule: lysyl-trna synthetase;PDBTitle: crystal structure of tetrameric form of human lysyl-trna2 synthetase
14 d1r6ra_
13.7
30
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C15 c1r6rA_
13.7
30
PDB header: viral proteinChain: A: PDB Molecule: genome polyprotein;PDBTitle: solution structure of dengue virus capsid protein reveals a2 new fold
16 c3c25A_
13.4
50
PDB header: hydrolase/dnaChain: A: PDB Molecule: noti restriction endonuclease;PDBTitle: crystal structure of noti restriction endonuclease bound to cognate2 dna
17 c1b8aB_
12.9
63
PDB header: ligaseChain: B: PDB Molecule: protein (aspartyl-trna synthetase);PDBTitle: aspartyl-trna synthetase
18 c1wydB_
12.6
63
PDB header: ligaseChain: B: PDB Molecule: hypothetical aspartyl-trna synthetase;PDBTitle: crystal structure of aspartyl-trna synthetase from sulfolobus tokodaii
19 c3fq6A_
12.5
42
PDB header: transferaseChain: A: PDB Molecule: methyltransferase;PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
20 c1x55A_
12.4
50
PDB header: ligaseChain: A: PDB Molecule: asparaginyl-trna synthetase;PDBTitle: crystal structure of asparaginyl-trna synthetase from pyrococcus2 horikoshii complexed with asparaginyl-adenylate analogue
21 c1e22A_
not modelled
12.3
50
PDB header: ligaseChain: A: PDB Molecule: lysyl-trna synthetase;PDBTitle: lysyl-trna synthetase (lysu) hexagonal form complexed with2 lysine and the non-hydrolysable atp analogue amp-pcp
22 c2xgtB_
not modelled
12.3
38
PDB header: ligaseChain: B: PDB Molecule: asparaginyl-trna synthetase, cytoplasmic;PDBTitle: asparaginyl-trna synthetase from brugia malayi complexed2 with the sulphamoyl analogue of asparaginyl-adenylate
23 c2kncA_
not modelled
11.8
0
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
24 d1o8bb1
not modelled
11.7
19
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain25 c1asyA_
not modelled
11.6
50
PDB header: complex (aminoacyl-trna synthase/trna)Chain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: class ii aminoacyl transfer rna synthetases: crystal2 structure of yeast aspartyl-trna synthetase complexed with3 trna asp
26 c3e9hB_
not modelled
11.0
50
PDB header: ligaseChain: B: PDB Molecule: lysyl-trna synthetase;PDBTitle: lysyl-trna synthetase from bacillus stearothermophilus2 complexed with l-lysylsulfamoyl adenosine
27 c3m4qA_
not modelled
10.8
38
PDB header: ligaseChain: A: PDB Molecule: asparaginyl-trna synthetase, putative;PDBTitle: entamoeba histolytica asparaginyl-trna synthetase (asnrs)
28 c1efwA_
not modelled
10.8
50
PDB header: ligase/rnaChain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: crystal structure of aspartyl-trna synthetase from thermus2 thermophilus complexed to trnaasp from escherichia coli
29 d2gf4a1
not modelled
10.2
17
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Vng1086c-likeFamily: Vng1086c-like30 c3mesB_
not modelled
10.2
31
PDB header: transferaseChain: B: PDB Molecule: choline kinase;PDBTitle: crystal structure of choline kinase from cryptosporidium2 parvum iowa ii, cgd3_2030
31 c1ceuA_
not modelled
9.9
25
PDB header: viral proteinChain: A: PDB Molecule: protein (hiv-1 regulatory protein n-terminalPDBTitle: nmr structure of the (1-51) n-terminal domain of the hiv-12 regulatory protein
32 c2voyK_
not modelled
9.7
14
PDB header: hydrolaseChain: K: PDB Molecule: sarcoplasmic/endoplasmic reticulum calciumPDBTitle: cryoem model of copa, the copper transporting atpase from2 archaeoglobus fulgidus
33 d1l0wa3
not modelled
9.7
50
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain34 d1c0aa3
not modelled
9.4
50
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain35 c2i3fA_
not modelled
9.4
13
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: glycolipid transfer-like protein;PDBTitle: crystal structure of a glycolipid transfer-like protein2 from galdieria sulphuraria
36 d3d37a2
not modelled
9.4
36
Fold: Phage tail proteinsSuperfamily: Phage tail proteinsFamily: Baseplate protein-like37 c1eqrC_
not modelled
9.2
50
PDB header: ligaseChain: C: PDB Molecule: aspartyl-trna synthetase;PDBTitle: crystal structure of free aspartyl-trna synthetase from2 escherichia coli
38 d1b8aa2
not modelled
9.2
63
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain39 d1eova2
not modelled
9.1
50
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain40 c3i7fA_
not modelled
9.1
38
PDB header: ligaseChain: A: PDB Molecule: aspartyl-trna synthetase;PDBTitle: aspartyl trna synthetase from entamoeba histolytica
41 c2ejbA_
not modelled
9.0
40
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: crystal structure of phenylacrylic acid decarboxylase from2 aquifex aeolicus
42 d1lgha_
not modelled
9.0
29
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits43 c2re3A_
not modelled
9.0
27
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf1285 family protein (spo_0140) from2 silicibacter pomeroyi dss-3 at 2.50 a resolution
44 c3rpfC_
not modelled
8.9
50
PDB header: transferaseChain: C: PDB Molecule: molybdopterin converting factor, subunit 1 (moad);PDBTitle: protein-protein complex of subunit 1 and 2 of molybdopterin-converting2 factor from helicobacter pylori 26695
45 c2hg5D_
not modelled
8.8
10
PDB header: membrane proteinChain: D: PDB Molecule: kcsa channel;PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
46 c3na2C_
not modelled
8.8
25
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function from mine drainage2 metagenome leptospirillum rubarum
47 d1sfka_
not modelled
8.7
19
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C48 d1sfkb_
not modelled
8.7
19
Fold: Flavivirus capsid protein CSuperfamily: Flavivirus capsid protein CFamily: Flavivirus capsid protein C49 c2uwjG_
not modelled
8.7
32
PDB header: chaperoneChain: G: PDB Molecule: type iii export protein pscg;PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
50 d1unca_
not modelled
8.6
33
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain51 c2jwaA_
not modelled
8.6
15
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
52 d1yu8x1
not modelled
8.5
33
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain53 c2p04B_
not modelled
8.4
14
PDB header: transferaseChain: B: PDB Molecule: signal transduction histidine kinase;PDBTitle: 2.1 ang structure of the dimerized pas domain of signal transduction2 histidine kinase from nostoc punctiforme pcc 73102 with homology to3 the h-noxa/h-noba domain of the soluble guanylyl cyclase
54 d1unda_
not modelled
8.3
33
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain55 c3g1zB_
not modelled
8.1
38
PDB header: ligaseChain: B: PDB Molecule: putative lysyl-trna synthetase;PDBTitle: structure of idp01693/yjea, a potential t-rna synthetase from2 salmonella typhimurium
56 c1m93A_
not modelled
8.1
15
PDB header: viral proteinChain: A: PDB Molecule: serine proteinase inhibitor 2;PDBTitle: 1.65 a structure of cleaved viral serpin crma
57 d1swxa_
not modelled
8.0
13
Fold: Glycolipid transfer protein, GLTPSuperfamily: Glycolipid transfer protein, GLTPFamily: Glycolipid transfer protein, GLTP58 d1sbza_
not modelled
7.9
40
Fold: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDSuperfamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCDFamily: Homo-oligomeric flavin-containing Cys decarboxylases, HFCD59 d2axth1
not modelled
7.9
10
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like60 d2axti1
not modelled
7.8
17
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein I, PsbIFamily: PsbI-like61 c2pjmA_
not modelled
7.8
19
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: structure of ribose 5-phosphate isomerase a from2 methanocaldococcus jannaschii
62 d1oqwa_
not modelled
7.8
20
Fold: Pili subunitsSuperfamily: Pili subunitsFamily: Pilin63 d1r89a1
not modelled
7.8
15
Fold: PAP/OAS1 substrate-binding domainSuperfamily: PAP/OAS1 substrate-binding domainFamily: Archaeal tRNA CCA-adding enzyme substrate-binding domain64 d2q7ra1
not modelled
7.6
21
Fold: MAPEG domain-likeSuperfamily: MAPEG domain-likeFamily: MAPEG domain65 d1nnha_
not modelled
7.5
38
Fold: Class II aaRS and biotin synthetasesSuperfamily: Class II aaRS and biotin synthetasesFamily: Class II aminoacyl-tRNA synthetase (aaRS)-like, catalytic domain66 d2b1xa1
not modelled
7.4
40
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain67 d2f05a1
not modelled
7.4
30
Fold: PAH2 domainSuperfamily: PAH2 domainFamily: PAH2 domain68 d1qzpa_
not modelled
7.4
21
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain69 c3kv0A_
not modelled
7.3
33
PDB header: transport proteinChain: A: PDB Molecule: het-c2;PDBTitle: crystal structure of het-c2: a fungal glycolipid transfer protein2 (gltp)
70 c2l3xA_
not modelled
7.3
21
PDB header: protein bindingChain: A: PDB Molecule: ablim2 protein;PDBTitle: villin head piece domain of human ablim2
71 d2qam31
not modelled
7.2
9
Fold: L35p-likeSuperfamily: L35p-likeFamily: Ribosomal protein L35p72 d1m0sa1
not modelled
7.2
45
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: D-ribose-5-phosphate isomerase (RpiA), catalytic domain73 c3zquA_
not modelled
7.2
30
PDB header: lyaseChain: A: PDB Molecule: probable aromatic acid decarboxylase;PDBTitle: structure of a probable aromatic acid decarboxylase
74 c2zylA_
not modelled
7.1
40
PDB header: oxidoreductaseChain: A: PDB Molecule: possible oxidoreductase;PDBTitle: crystal structure of 3-ketosteroid-9-alpha-hydroxylase2 (ksha) from m. tuberculosis
75 d1zvpa1
not modelled
7.0
22
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: VC0802-like76 c2k6nA_
not modelled
7.0
22
PDB header: structural proteinChain: A: PDB Molecule: supervillin;PDBTitle: solution structure of human supervillin headpiece, minimized2 average
77 c2qieB_
not modelled
7.0
50
PDB header: transferaseChain: B: PDB Molecule: molybdopterin synthase small subunit;PDBTitle: staphylococcus aureus molybdopterin synthase in complex2 with precursor z
78 c3cu4A_
not modelled
6.9
17
PDB header: electron transportChain: A: PDB Molecule: cytochrome c family protein;PDBTitle: omcf, outer membrance cytochrome f from geobacter2 sulfurreducens
79 d2cosa1
not modelled
6.9
17
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain80 c1m0sA_
not modelled
6.9
28
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: northeast structural genomics consortium (nesg id ir21)
81 c1lkzB_
not modelled
6.9
19
PDB header: isomeraseChain: B: PDB Molecule: ribose 5-phosphate isomerase a;PDBTitle: crystal structure of d-ribose-5-phosphate isomerase (rpia)2 from escherichia coli.
82 c2kwtA_
not modelled
6.9
27
PDB header: viral proteinChain: A: PDB Molecule: protease ns2-3;PDBTitle: solution structure of ns2 [27-59]
83 d1jhda1
not modelled
6.9
17
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: ATP sulfurylase N-terminal domain84 d1yu5x1
not modelled
6.8
33
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain85 c3bs7A_
not modelled
6.8
13
PDB header: signaling proteinChain: A: PDB Molecule: protein aveugle;PDBTitle: crystal structure of the sterile alpha motif (sam) domain2 of hyphen/aveugle
86 d1p9qc3
not modelled
6.8
33
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain87 d1o7na1
not modelled
6.8
20
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain88 d1ulia1
not modelled
6.7
20
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain89 d1wqla1
not modelled
6.7
20
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain90 d1fm0d_
not modelled
6.7
50
Fold: beta-Grasp (ubiquitin-like)Superfamily: MoaD/ThiSFamily: MoaD91 d2ha9a1
not modelled
6.7
23
Fold: PFL-like glycyl radical enzymesSuperfamily: PFL-like glycyl radical enzymesFamily: SP0239-like92 c3gteB_
not modelled
6.7
20
PDB header: electron transport, oxidoreductaseChain: B: PDB Molecule: ddmc;PDBTitle: crystal structure of dicamba monooxygenase with non-heme2 iron
93 c2q1kA_
not modelled
6.6
44
PDB header: chaperoneChain: A: PDB Molecule: asce;PDBTitle: cyrstal structure of asce from aeromonas hydrophilla
94 d1gdva_
not modelled
6.6
20
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: monodomain cytochrome c95 d1ujsa_
not modelled
6.6
11
Fold: VHP, Villin headpiece domainSuperfamily: VHP, Villin headpiece domainFamily: VHP, Villin headpiece domain96 c3acgA_
not modelled
6.6
50
PDB header: hydrolaseChain: A: PDB Molecule: beta-1,4-endoglucanase;PDBTitle: crystal structure of carbohydrate-binding module family 282 from clostridium josui cel5a in complex with cellobiose
97 c1w2lA_
not modelled
6.6
17
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome oxidase subunit ii;PDBTitle: cytochrome c domain of caa3 oxygen oxidoreductase
98 c3gkqB_
not modelled
6.6
20
PDB header: oxidoreductaseChain: B: PDB Molecule: terminal oxygenase component of carbazole 1,9a-PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 novosphingobium sp. ka1
99 d1vzva_
not modelled
6.5
29
Fold: Herpes virus serine proteinase, assemblinSuperfamily: Herpes virus serine proteinase, assemblinFamily: Herpes virus serine proteinase, assemblin