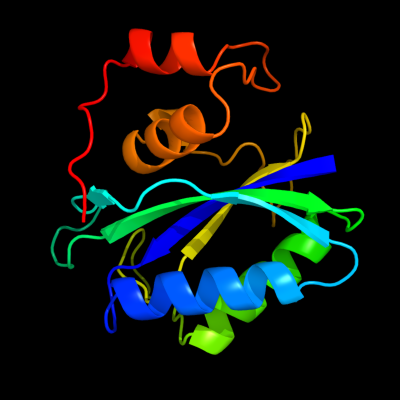
| 1 | d1f9ya_
|
|
 |
100.0 |
100 |
Fold:Ferredoxin-like
Superfamily:6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPK
Family:6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPK |
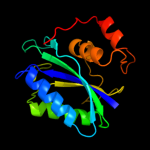
| 2 | d1cbka_
|
|
 |
100.0 |
57 |
Fold:Ferredoxin-like
Superfamily:6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPK
Family:6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase, HPPK |
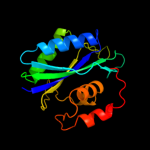
| 3 | c2qx0A_
|
|
 |
100.0 |
63 |
PDB header:transferase
Chain: A: PDB Molecule:7,8-dihydro-6-hydroxymethylpterin-
PDBTitle: crystal structure of yersinia pestis hppk (ternary complex)
|
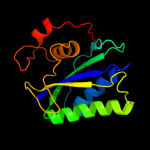
| 4 | c2bmbA_
|
|
 |
100.0 |
33 |
PDB header:transferase
Chain: A: PDB Molecule:folic acid synthesis protein fol1;
PDBTitle: x-ray structure of the bifunctional 6-hydroxymethyl-7,8-2 dihydroxypterin pyrophosphokinase dihydropteroate synthase3 from saccharomyces cerevisiae
|
| 5 | c3mcnA_
|
|
 |
100.0 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:2-amino-4-hydroxy-6-hydroxymethyldihydropteridine
PDBTitle: crystal structure of the 6-hyroxymethyl-7,8-dihydropterin2 pyrophosphokinase dihydropteroate synthase bifunctional enzyme from3 francisella tularensis
|
| 6 | c2cg8B_
|
|
 |
100.0 |
34 |
PDB header:lyase/transferase
Chain: B: PDB Molecule:dihydroneopterin aldolase 6-hydroxymethyl-7,8-
PDBTitle: the bifunctional dihydroneopterin aldolase 6-hydroxymethyl-2 7,8-dihydropterin synthase from streptococcus pneumoniae
|
| 7 | c3r3tA_
|
|
 |
31.1 |
15 |
PDB header:rna binding protein
Chain: A: PDB Molecule:30s ribosomal protein s6;
PDBTitle: crystal structure of 30s ribosomal protein s from bacillus anthracis
|
| 8 | d2j5aa1
|
|
 |
28.1 |
28 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 9 | c2o6lA_
|
|
 |
24.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glucuronosyltransferase 2b7;
PDBTitle: crystal structure of the udp-glucuronic acid binding domain2 of the human drug metabolizing udp-glucuronosyltransferase3 2b7
|
| 10 | c3ndcB_
|
|
 |
23.0 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
| 11 | d1qjha_
|
|
 |
22.8 |
23 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 12 | d1loua_
|
|
 |
20.0 |
26 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 13 | c3bbnF_
|
|
 |
15.0 |
19 |
PDB header:ribosome
Chain: F: PDB Molecule:ribosomal protein s6;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 14 | c3idwA_
|
|
 |
13.7 |
13 |
PDB header:endocytosis
Chain: A: PDB Molecule:actin cytoskeleton-regulatory complex protein sla1;
PDBTitle: crystal structure of sla1 homology domain 2
|
| 15 | c2inpD_
|
|
 |
13.4 |
19 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:phenol hydroxylase component phl;
PDBTitle: structure of the phenol hydroxylase-regulatory protein2 complex
|
| 16 | d1t6ca1
|
|
 |
12.3 |
64 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
| 17 | d1vmba_
|
|
 |
12.3 |
24 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 18 | c1vmbA_
|
|
 |
12.3 |
24 |
PDB header:translation
Chain: A: PDB Molecule:30s ribosomal protein s6;
PDBTitle: crystal structure of 30s ribosomal protein s6 (tm0603) from thermotoga2 maritima at 1.70 a resolution
|
| 19 | d2bcgg3
|
|
 |
11.8 |
29 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GDI-like |
| 20 | d1d5ta2
|
|
 |
10.9 |
34 |
Fold:FAD-linked reductases, C-terminal domain
Superfamily:FAD-linked reductases, C-terminal domain
Family:GDI-like |
| 21 | c1u9gA_ |
|
not modelled |
10.9 |
37 |
PDB header:transcription
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(8)l(9)
|
| 22 | c2wc1A_ |
|
not modelled |
10.0 |
15 |
PDB header:electron transport
Chain: A: PDB Molecule:flavodoxin;
PDBTitle: three-dimensional structure of the nitrogen fixation2 flavodoxin (niff) from rhodobacter capsulatus at 2.2 a
|
| 23 | c1w5kD_ |
|
not modelled |
9.6 |
32 |
PDB header:four helix bundle
Chain: D: PDB Molecule:general control protein gcn4;
PDBTitle: an anti-parallel four helix bundle
|
| 24 | c1w5kC_ |
|
not modelled |
9.6 |
32 |
PDB header:four helix bundle
Chain: C: PDB Molecule:general control protein gcn4;
PDBTitle: an anti-parallel four helix bundle
|
| 25 | c1w5kB_ |
|
not modelled |
9.6 |
32 |
PDB header:four helix bundle
Chain: B: PDB Molecule:general control protein gcn4;
PDBTitle: an anti-parallel four helix bundle
|
| 26 | c1w5kA_ |
|
not modelled |
9.6 |
32 |
PDB header:four helix bundle
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: an anti-parallel four helix bundle
|
| 27 | c1yzvA_ |
|
not modelled |
9.4 |
7 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: hypothetical protein from trypanosoma cruzi
|
| 28 | c1unyA_ |
|
not modelled |
8.7 |
38 |
PDB header:four helix bundle
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
|
| 29 | c1unvB_ |
|
not modelled |
8.6 |
38 |
PDB header:four helix bundle
Chain: B: PDB Molecule:general control protein gcn4;
PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
|
| 30 | c1unvA_ |
|
not modelled |
8.5 |
38 |
PDB header:four helix bundle
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
|
| 31 | c2ehbD_ |
|
not modelled |
8.3 |
14 |
PDB header:signalling protein/transferase
Chain: D: PDB Molecule:cbl-interacting serine/threonine-protein kinase 24;
PDBTitle: the structure of the c-terminal domain of the protein kinase atsos22 bound to the calcium sensor atsos3
|
| 32 | c2q4dB_ |
|
not modelled |
8.2 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:lysine decarboxylase-like protein at5g11950;
PDBTitle: ensemble refinement of the crystal structure of a lysine2 decarboxylase-like protein from arabidopsis thaliana gene at5g11950
|
| 33 | c2kjwA_ |
|
not modelled |
7.9 |
26 |
PDB header:ribosomal protein
Chain: A: PDB Molecule:30s ribosomal protein s6;
PDBTitle: solution structure and backbone dynamics of the permutant2 p54-55
|
| 34 | c1u9gB_ |
|
not modelled |
7.9 |
38 |
PDB header:transcription
Chain: B: PDB Molecule:general control protein gcn4;
PDBTitle: heterocyclic peptide backbone modification in gcn4-pli based coiled2 coils: replacement of k(8)l(9)
|
| 35 | c2la3A_ |
|
not modelled |
7.8 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: the nmr structure of the protein np_344798.1 reveals a cca-adding2 enzyme head domain
|
| 36 | c1unzA_ |
|
not modelled |
7.5 |
38 |
PDB header:four helix bundle
Chain: A: PDB Molecule:general control protein gcn4;
PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
|
| 37 | c1unzB_ |
|
not modelled |
7.5 |
38 |
PDB header:four helix bundle
Chain: B: PDB Molecule:general control protein gcn4;
PDBTitle: structure based engineering of internal molecular surfaces2 of four helix bundles
|
| 38 | c3a4cA_ |
|
not modelled |
7.5 |
33 |
PDB header:cell cycle, replication
Chain: A: PDB Molecule:dna replication factor cdt1;
PDBTitle: crystal structure of cdt1 c terminal domain
|
| 39 | c2qikA_ |
|
not modelled |
7.1 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0131 protein ykqa;
PDBTitle: crystal structure of ykqa from bacillus subtilis. northeast2 structural genomics target sr631
|
| 40 | c2qbbF_ |
|
not modelled |
7.0 |
19 |
PDB header:ribosome
Chain: F: PDB Molecule:30s ribosomal protein s6;
PDBTitle: crystal structure of the bacterial ribosome from2 escherichia coli in complex with gentamicin. this file3 contains the 30s subunit of the second 70s ribosome, with4 gentamicin bound. the entire crystal structure contains5 two 70s ribosomes and is described in remark 400.
|
| 41 | d2qalf1 |
|
not modelled |
7.0 |
19 |
Fold:Ferredoxin-like
Superfamily:Ribosomal protein S6
Family:Ribosomal protein S6 |
| 42 | d1dd3a1 |
|
not modelled |
7.0 |
26 |
Fold:Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily:Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family:Ribosomal protein L7/12, oligomerisation (N-terminal) domain |
| 43 | c2zdiC_ |
|
not modelled |
6.6 |
19 |
PDB header:chaperone
Chain: C: PDB Molecule:prefoldin subunit alpha;
PDBTitle: crystal structure of prefoldin from pyrococcus horikoshii2 ot3
|
| 44 | c3s7xC_ |
|
not modelled |
6.5 |
25 |
PDB header:viral protein
Chain: C: PDB Molecule:major capsid protein vp1;
PDBTitle: unassembled washington university polyomavirus vp1 pentamer r198k2 mutant
|
| 45 | c2dboA_ |
|
not modelled |
6.3 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:d-tyrosyl-trna(tyr) deacylase;
PDBTitle: crystal structure of d-tyr-trna(tyr) deacylase from aquifex aeolicus
|
| 46 | d1tjna_ |
|
not modelled |
5.9 |
16 |
Fold:Chelatase-like
Superfamily:Chelatase
Family:CbiX-like |
| 47 | c1tjnA_ |
|
not modelled |
5.9 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:sirohydrochlorin cobaltochelatase;
PDBTitle: crystal structure of hypothetical protein af0721 from archaeoglobus2 fulgidus
|
| 48 | c1zawV_ |
|
not modelled |
5.8 |
26 |
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
|
| 49 | c1zawU_ |
|
not modelled |
5.8 |
26 |
PDB header:structural protein
Chain: U: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
|
| 50 | c1zawW_ |
|
not modelled |
5.8 |
26 |
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
|
| 51 | c1zaxV_ |
|
not modelled |
5.6 |
26 |
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
|
| 52 | c1zavW_ |
|
not modelled |
5.6 |
26 |
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
|
| 53 | c1zaxW_ |
|
not modelled |
5.6 |
26 |
PDB header:structural protein
Chain: W: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
|
| 54 | c1zavV_ |
|
not modelled |
5.6 |
26 |
PDB header:structural protein
Chain: V: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
|
| 55 | d1hn0a4 |
|
not modelled |
5.6 |
56 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Hyaluronate lyase-like, central domain |
| 56 | d1fxkc_ |
|
not modelled |
5.6 |
8 |
Fold:Long alpha-hairpin
Superfamily:Prefoldin
Family:Prefoldin |
| 57 | c1dd3C_ |
|
not modelled |
5.4 |
26 |
PDB header:ribosome
Chain: C: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
|
| 58 | c1dd3D_ |
|
not modelled |
5.4 |
26 |
PDB header:ribosome
Chain: D: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
|
| 59 | d1dd4c_ |
|
not modelled |
5.3 |
26 |
Fold:Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Superfamily:Ribosomal protein L7/12, oligomerisation (N-terminal) domain
Family:Ribosomal protein L7/12, oligomerisation (N-terminal) domain |
| 60 | c3g9rF_ |
|
not modelled |
5.3 |
42 |
PDB header:viral protein
Chain: F: PDB Molecule:fusion complex of hiv-1 envelope glycoprotein
PDBTitle: structure of the hiv-1 gp41 membrane-proximal ectodomain2 region in a putative prefusion conformation
|
| 61 | c2rbhA_ |
|
not modelled |
5.3 |
38 |
PDB header:transferase
Chain: A: PDB Molecule:gamma-glutamyl cyclotransferase;
PDBTitle: gamma-glutamyl cyclotransferase
|
| 62 | c1zaxZ_ |
|
not modelled |
5.1 |
28 |
PDB header:structural protein
Chain: Z: PDB Molecule:50s ribosomal protein l7/l12;
PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
|