| 1 |
|
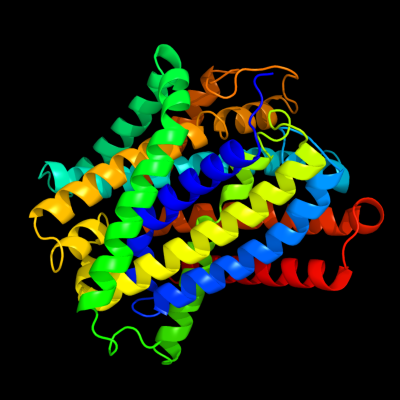
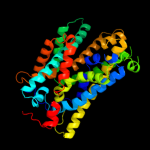
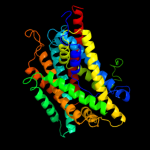
PDB 3gia chain A
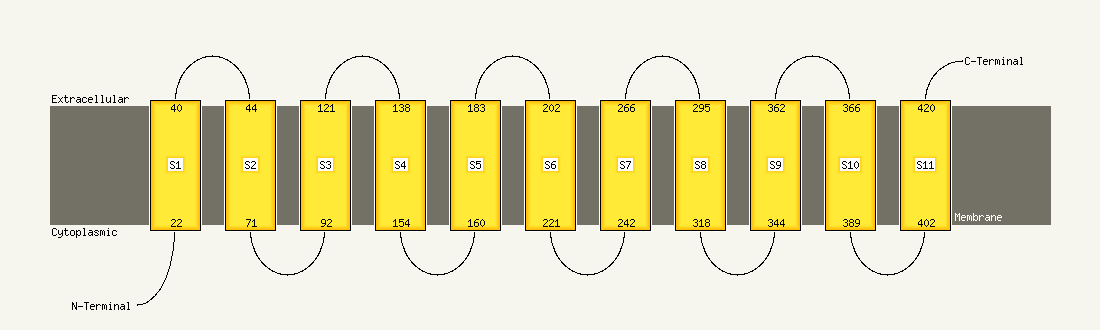
Region: 14 - 420
Aligned: 392
Modelled: 392
Confidence: 100.0%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
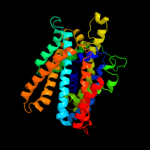
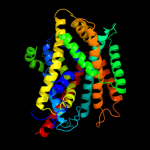
PDB 3lrc chain C
Region: 14 - 419
Aligned: 370
Modelled: 370
Confidence: 100.0%
Identity: 13%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
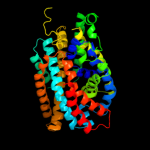
PDB 2jln chain A
Region: 4 - 399
Aligned: 375
Modelled: 375
Confidence: 99.9%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
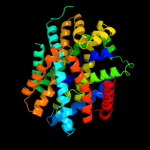
PDB 2xq2 chain A
Region: 15 - 419
Aligned: 393
Modelled: 405
Confidence: 99.0%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 15 - 419
Aligned: 394
Modelled: 394
Confidence: 98.9%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 17 - 420
Aligned: 395
Modelled: 395
Confidence: 98.5%
Identity: 16%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 10 - 399
Aligned: 375
Modelled: 382
Confidence: 98.3%
Identity: 8%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 5 - 397
Aligned: 384
Modelled: 384
Confidence: 95.9%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2hg5 chain D
Region: 18 - 48
Aligned: 31
Modelled: 31
Confidence: 10.0%
Identity: 10%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 10 |
|
PDB 3klz chain E
Region: 10 - 38
Aligned: 29
Modelled: 29
Confidence: 6.6%
Identity: 14%
PDB header:membrane protein
Chain: E: PDB Molecule:putative formate transporter 1;
PDBTitle: pentameric formate channel with formate bound
Phyre2
| 11 |
|
PDB 3mp7 chain B
Region: 386 - 422
Aligned: 37
Modelled: 37
Confidence: 5.9%
Identity: 22%
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
Phyre2