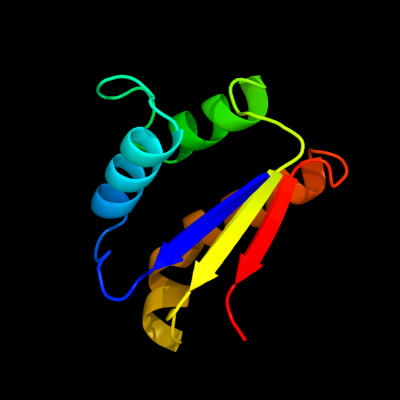
| 1 | d2cx6a1
|
|
 |
100.0 |
98 |
Fold:Barstar-like
Superfamily:Barstar-related
Family:Barstar-related |
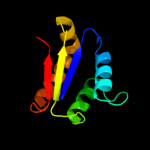
| 2 | d1ay7b_
|
|
 |
99.9 |
24 |
Fold:Barstar-like
Superfamily:Barstar-related
Family:Barstar-related |
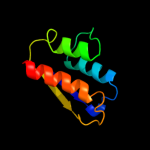
| 3 | c1sb7A_
|
|
 |
68.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:trna pseudouridine synthase d;
PDBTitle: crystal structure of the e.coli pseudouridine synthase trud
|
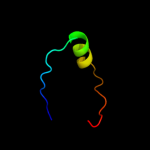
| 4 | d1szwa_
|
|
 |
68.2 |
16 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:tRNA pseudouridine synthase TruD |
| 5 | c3nmeA_
|
|
 |
24.0 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:sex4 glucan phosphatase;
PDBTitle: structure of a plant phosphatase
|
| 6 | d2ahob1
|
|
 |
23.6 |
6 |
Fold:SAM domain-like
Superfamily:eIF2alpha middle domain-like
Family:eIF2alpha middle domain-like |
| 7 | d1nksa_
|
|
 |
21.2 |
7 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 8 | c2c0kB_
|
|
 |
18.9 |
10 |
PDB header:oxygen transport
Chain: B: PDB Molecule:hemoglobin;
PDBTitle: the structure of hemoglobin from the botfly gasterophilus2 intestinalis
|
| 9 | c1z2zB_
|
|
 |
18.5 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:probable trna pseudouridine synthase d;
PDBTitle: crystal structure of the putative trna pseudouridine2 synthase d (trud) from methanosarcina mazei, northeast3 structural genomics target mar1
|
| 10 | c3da5A_
|
|
 |
18.5 |
45 |
PDB header:rna binding protein
Chain: A: PDB Molecule:argonaute;
PDBTitle: crystal structure of piwi/argonaute/zwille(paz) domain from2 thermococcus thioreducens
|
| 11 | d1vpya_
|
|
 |
17.9 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:TM1631-like
Family:TM1631-like |
| 12 | c2dnfA_
|
|
 |
17.4 |
4 |
PDB header:protein binding
Chain: A: PDB Molecule:doublecortin domain-containing protein 2;
PDBTitle: solution structure of rsgi ruh-062, a dcx domain from human
|
| 13 | c3f1xA_
|
|
 |
16.7 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:serine acetyltransferase;
PDBTitle: three dimensional structure of the serine acetyltransferase from2 bacteroides vulgatus, northeast structural genomics consortium target3 bvr62.
|
| 14 | c2zpmA_
|
|
 |
14.5 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
|
| 15 | d2hgaa1
|
|
 |
12.7 |
5 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:MTH1368 C-terminal domain-like |
| 16 | c2k5cA_
|
|
 |
12.5 |
16 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein pf0385;
PDBTitle: nmr structure for pf0385
|
| 17 | d1gg3a3
|
|
 |
12.2 |
10 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:First domain of FERM |
| 18 | c2grjH_
|
|
 |
11.7 |
4 |
PDB header:transferase
Chain: H: PDB Molecule:dephospho-coa kinase;
PDBTitle: crystal structure of dephospho-coa kinase (ec 2.7.1.24)2 (dephosphocoenzyme a kinase) (tm1387) from thermotoga maritima at3 2.60 a resolution
|
| 19 | d1y14b2
|
|
 |
11.6 |
0 |
Fold:Dodecin subunit-like
Superfamily:N-terminal, heterodimerisation domain of RBP7 (RpoE)
Family:N-terminal, heterodimerisation domain of RBP7 (RpoE) |
| 20 | d2c4va1
|
|
 |
10.4 |
15 |
Fold:Flavodoxin-like
Superfamily:Type II 3-dehydroquinate dehydratase
Family:Type II 3-dehydroquinate dehydratase |
| 21 | d1sota1 |
|
not modelled |
9.6 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 22 | d1ky9b2 |
|
not modelled |
9.5 |
5 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 23 | d1wuda1 |
|
not modelled |
9.3 |
11 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:HRDC domain from helicases |
| 24 | d2ciwa2 |
|
not modelled |
9.1 |
11 |
Fold:EF Hand-like
Superfamily:Cloroperoxidase
Family:Cloroperoxidase |
| 25 | c3q3uA_ |
|
not modelled |
9.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lignin peroxidase;
PDBTitle: trametes cervina lignin peroxidase
|
| 26 | d2zpya3 |
|
not modelled |
8.9 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:First domain of FERM |
| 27 | c2p3wB_ |
|
not modelled |
8.4 |
5 |
PDB header:protein binding
Chain: B: PDB Molecule:probable serine protease htra3;
PDBTitle: crystal structure of the htra3 pdz domain bound to a phage-derived2 ligand (fgrwv)
|
| 28 | c2xxsA_ |
|
not modelled |
8.4 |
11 |
PDB header:protein binding
Chain: A: PDB Molecule:protein mxig;
PDBTitle: solution structure of the n-terminal domain of the shigella2 type iii secretion protein mxig
|
| 29 | c3i18A_ |
|
not modelled |
7.6 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lmo2051 protein;
PDBTitle: crystal structure of the pdz domain of the sdrc-like protein2 (lmo2051) from listeria monocytogenes, northeast structural3 genomics consortium target lmr166b
|
| 30 | d2e1fa1 |
|
not modelled |
7.6 |
6 |
Fold:SAM domain-like
Superfamily:HRDC-like
Family:HRDC domain from helicases |
| 31 | c2kl1A_ |
|
not modelled |
7.5 |
19 |
PDB header:protein binding
Chain: A: PDB Molecule:ylbl protein;
PDBTitle: solution structure of gtr34c from geobacillus thermodenitrificans.2 northeast structural genomics consortium target gtr34c
|
| 32 | c2x7jA_ |
|
not modelled |
7.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:2-succinyl-5-enolpyruvyl-6-hydroxy-3-cyclohexene
PDBTitle: structure of the menaquinone biosynthesis protein mend from2 bacillus subtilis
|
| 33 | d1ni2a3 |
|
not modelled |
7.1 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:First domain of FERM |
| 34 | d2z9ia1 |
|
not modelled |
6.9 |
14 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 35 | c2o42B_ |
|
not modelled |
6.9 |
19 |
PDB header:apoptosis inhibitor
Chain: B: PDB Molecule:m11l protein;
PDBTitle: crystal structure of m11l, bcl-2 homolog from myxoma virus
|
| 36 | c3bq9A_ |
|
not modelled |
6.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted rossmann fold nucleotide-binding domain-
PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
|
| 37 | d1itha_ |
|
not modelled |
6.6 |
15 |
Fold:Globin-like
Superfamily:Globin-like
Family:Globins |
| 38 | d1ef1a3 |
|
not modelled |
6.6 |
13 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:First domain of FERM |
| 39 | c1yz7A_ |
|
not modelled |
6.6 |
9 |
PDB header:translation
Chain: A: PDB Molecule:probable translation initiation factor 2 alpha
PDBTitle: crystal structure of a c-terminal segment of the alpha2 subunit of aif2 from pyrococcus abyssi
|
| 40 | d1sxja1 |
|
not modelled |
6.5 |
0 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:DNA polymerase III clamp loader subunits, C-terminal domain |
| 41 | d1ukza_ |
|
not modelled |
6.5 |
0 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nucleotide and nucleoside kinases |
| 42 | d2r8oa2 |
|
not modelled |
6.5 |
15 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:TK-like PP module |
| 43 | c3u8zD_ |
|
not modelled |
5.8 |
17 |
PDB header:signaling protein
Chain: D: PDB Molecule:merlin;
PDBTitle: human merlin ferm domain
|
| 44 | c2joaA_ |
|
not modelled |
5.8 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:serine protease htra1;
PDBTitle: htra1 bound to an optimized peptide: nmr assignment of pdz2 domain and ligand resonances
|
| 45 | c2q9rA_ |
|
not modelled |
5.7 |
26 |
PDB header:unknown function
Chain: A: PDB Molecule:protein of unknown function;
PDBTitle: crystal structure of a duf416 family protein (sbal_3149) from2 shewanella baltica os155 at 1.91 a resolution
|
| 46 | d1yyda1 |
|
not modelled |
5.5 |
17 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:CCP-like |
| 47 | d2e39a1 |
|
not modelled |
5.5 |
22 |
Fold:Heme-dependent peroxidases
Superfamily:Heme-dependent peroxidases
Family:CCP-like |
| 48 | d1tw9a2 |
|
not modelled |
5.4 |
20 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 49 | d1orjb_ |
|
not modelled |
5.2 |
18 |
Fold:Four-helical up-and-down bundle
Superfamily:Flagellar export chaperone FliS
Family:Flagellar export chaperone FliS |
| 50 | d2i6va1 |
|
not modelled |
5.2 |
5 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:EpsC C-terminal domain-like |
| 51 | d3c8ya2 |
|
not modelled |
5.1 |
8 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:2Fe-2S ferredoxin-like
Family:2Fe-2S ferredoxin domains from multidomain proteins |
| 52 | c2vvyC_ |
|
not modelled |
5.1 |
32 |
PDB header:viral protein
Chain: C: PDB Molecule:protein b15;
PDBTitle: structure of vaccinia virus protein b14
|
| 53 | d1glqa2 |
|
not modelled |
5.0 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 54 | c2z5sN_ |
|
not modelled |
5.0 |
8 |
PDB header:cell cycle
Chain: N: PDB Molecule:mdm4 protein;
PDBTitle: molecular basis for the inhibition of p53 by mdmx
|