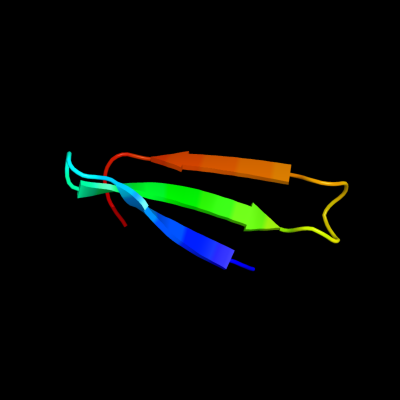
1 c2yztA_
35.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein ttha1756;PDBTitle: crystal structure of uncharacterized conserved protein from thermus2 thermophilus hb8
2 d2j8ka1
31.0
14
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats3 c3pssB_
28.3
38
PDB header: cell cycleChain: B: PDB Molecule: qnr;PDBTitle: crystal structure of ahqnr, the qnr protein from aeromonas hydrophila2 (p21 crystal form)
4 c2g0yA_
28.1
27
PDB header: unknown functionChain: A: PDB Molecule: pentapeptide repeat protein;PDBTitle: crystal structure of a lumenal pentapeptide repeat protein from2 cyanothece sp 51142 at 2.3 angstrom resolution. tetragonal crystal3 form
5 c3du1X_
24.2
12
PDB header: structural proteinChain: X: PDB Molecule: all3740 protein;PDBTitle: the 2.0 angstrom resolution crystal structure of hetl, a pentapeptide2 repeat protein involved in heterocyst differentiation regulation from3 the cyanobacterium nostoc sp. strain pcc 7120
6 c3n90A_
20.2
31
PDB header: unknown functionChain: A: PDB Molecule: thylakoid lumenal 15 kda protein 1, chloroplastic;PDBTitle: the 1.7 angstrom resolution crystal structure of at2g44920, a2 pentapeptide repeat protein from arabidopsis thaliana thylakoid3 lumen.
7 c2vy2A_
20.2
41
PDB header: transcriptionChain: A: PDB Molecule: protein leafy;PDBTitle: structure of leafy transcription factor from arabidopsis2 thaliana in complex with dna from ag-i promoter
8 d2j8ia1
19.9
13
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats9 c2xt4A_
19.4
21
PDB header: cell cycleChain: A: PDB Molecule: mcbg-like protein;PDBTitle: structure of the pentapeptide repeat protein albg, a2 resistance factor for the topoisomerase poison albicidin.
10 c3cucB_
18.9
24
PDB header: signaling proteinChain: B: PDB Molecule: protein of unknown function with a fic domain;PDBTitle: crystal structure of a fic domain containing signaling protein2 (bt_2513) from bacteroides thetaiotaomicron vpi-5482 at 2.71 a3 resolution
11 c3k1iA_
18.4
16
PDB header: chaperoneChain: A: PDB Molecule: flagellar protein;PDBTitle: crystal strcture of flis-hp1076 complex in h. pylori
12 c2xtwB_
17.8
17
PDB header: cell cycleChain: B: PDB Molecule: qnrb1;PDBTitle: structure of qnrb1 (full length), a plasmid-mediated2 fluoroquinolone resistance protein
13 c2xt4B_
15.0
18
PDB header: cell cycleChain: B: PDB Molecule: mcbg-like protein;PDBTitle: structure of the pentapeptide repeat protein albg, a2 resistance factor for the topoisomerase poison albicidin.
14 d1u4ra_
14.5
33
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines15 c3oiqB_
13.7
89
PDB header: protein bindingChain: B: PDB Molecule: dna polymerase alpha catalytic subunit a;PDBTitle: crystal structure of yeast telomere protein cdc13 ob1 and the2 catalytic subunit of dna polymerase alpha pol1
16 c2w7zB_
13.6
18
PDB header: inhibitorChain: B: PDB Molecule: pentapeptide repeat family protein;PDBTitle: structure of the pentapeptide repeat protein efsqnr, a dna2 gyrase inhibitor. free amines modified by cyclic3 pentylation with glutaraldehyde.
17 c3gb8B_
13.4
18
PDB header: transport proteinChain: B: PDB Molecule: snurportin-1;PDBTitle: crystal structure of crm1/snurportin-1 complex
18 d1m8aa_
13.4
21
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines19 c2j8kA_
12.9
18
PDB header: toxinChain: A: PDB Molecule: np275-np276;PDBTitle: structure of the fusion of np275 and np276, pentapeptide2 repeat proteins from nostoc punctiforme
20 d3lkfa_
11.9
21
Fold: Leukocidin-likeSuperfamily: Leukocidin-likeFamily: Leukocidin (pore-forming toxin)21 d2hcca_
not modelled
11.8
28
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines22 c3h6sE_
not modelled
11.6
63
PDB header: hydrolase/hydrolase inhibitorChain: E: PDB Molecule: clitocypin analog;PDBTitle: strucure of clitocypin - cathepsin v complex
23 d1g91a_
not modelled
11.2
39
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines24 d1ex0a2
not modelled
10.9
17
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Transglutaminase, two C-terminal domainsFamily: Transglutaminase, two C-terminal domains25 d2g03a1
not modelled
10.9
13
Fold: Fic-likeSuperfamily: Fic-likeFamily: Fic-like26 c3oc5A_
not modelled
10.4
36
PDB header: cell adhesionChain: A: PDB Molecule: toxin coregulated pilus biosynthesis protein f;PDBTitle: crystal structure of the vibrio cholerae secreted colonization factor2 tcpf
27 d1j8ia_
not modelled
10.3
22
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines28 c2l4nA_
not modelled
10.1
39
PDB header: cytokineChain: A: PDB Molecule: c-c motif chemokine 21;PDBTitle: solution structure of the chemokine ccl21
29 d2eota_
not modelled
10.1
33
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines30 c2o6wA_
not modelled
10.0
15
PDB header: unknown functionChain: A: PDB Molecule: repeat five residue (rfr) protein orPDBTitle: crystal structure of a pentapeptide repeat protein (rfr23)2 from the cyanobacterium cyanothece 51142
31 d2f3la1
not modelled
9.5
28
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats32 d1bo0a_
not modelled
9.4
39
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines33 c2ra4B_
not modelled
9.4
28
PDB header: cytokineChain: B: PDB Molecule: small-inducible cytokine a13;PDBTitle: crystal structure of human monocyte chemoattractant protein 4 (mcp-2 4/ccl13)
34 d1huna_
not modelled
9.1
28
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines35 d1eiga_
not modelled
8.9
33
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines36 d1c8za_
not modelled
8.7
67
Fold: Tubby C-terminal domain-likeSuperfamily: Tubby C-terminal domain-likeFamily: Transcriptional factor tubby, C-terminal domain37 d2bm5a1
not modelled
8.1
24
Fold: Single-stranded right-handed beta-helixSuperfamily: Pentapeptide repeat-likeFamily: Pentapeptide repeats38 c2j8iB_
not modelled
7.9
16
PDB header: toxinChain: B: PDB Molecule: np275;PDBTitle: structure of np275, a pentapeptide repeat protein from2 nostoc punctiforme
39 d1el0a_
not modelled
7.9
33
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines40 d1nr4a_
not modelled
7.9
28
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines41 c3bq9A_
not modelled
7.8
32
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: predicted rossmann fold nucleotide-binding domain-PDBTitle: crystal structure of predicted nucleotide-binding protein from2 idiomarina baltica os145
42 d1nr4b_
not modelled
7.7
28
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines43 d1b50a_
not modelled
7.5
39
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines44 d1n28a_
not modelled
7.4
22
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A245 d1doka_
not modelled
7.2
39
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines46 d1iaza_
not modelled
7.1
21
Fold: Cytolysin/lectinSuperfamily: Cytolysin/lectinFamily: Anemone pore-forming cytolysin47 d1tg7a4
not modelled
7.0
27
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: Beta-galactosidase LacA, domain 248 d1eaqa_
not modelled
7.0
35
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: RUNT domain49 d2pbea2
not modelled
7.0
50
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: AadK N-terminal domain-like50 d1jltb_
not modelled
6.9
20
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A251 d1g2ta_
not modelled
6.9
28
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines52 c2q8rF_
not modelled
6.7
28
PDB header: cytokineChain: F: PDB Molecule: ccl14;PDBTitle: structural and functional characterization of cc chemokine ccl14
53 c1zxtD_
not modelled
6.7
28
PDB header: signaling proteinChain: D: PDB Molecule: functional macrophage inflammatory protein 1-alpha homolog;PDBTitle: crystal structure of a viral chemokine
54 c2cazF_
not modelled
6.6
31
PDB header: protein transportChain: F: PDB Molecule: protein srn2;PDBTitle: escrt-i core
55 d1b3aa_
not modelled
6.6
33
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines56 d1ddba_
not modelled
6.5
27
Fold: Toxins' membrane translocation domainsSuperfamily: Bcl-2 inhibitors of programmed cell deathFamily: Bcl-2 inhibitors of programmed cell death57 c3n3vA_
not modelled
6.4
14
PDB header: transferaseChain: A: PDB Molecule: adenosine monophosphate-protein transferase ibpa;PDBTitle: crystal structure of ibpafic2-h3717a in complex with adenylylated2 cdc42
58 d2nz1d1
not modelled
6.2
39
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines59 d1ljma_
not modelled
6.2
35
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: p53-like transcription factorsFamily: RUNT domain60 d1gmza_
not modelled
5.8
17
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A261 d1nxza1
not modelled
5.5
23
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: YggJ N-terminal domain-like62 d1g2xa_
not modelled
5.5
26
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A263 d2fhta1
not modelled
5.3
33
Fold: IL8-likeSuperfamily: Interleukin 8-like chemokinesFamily: Interleukin 8-like chemokines64 d1oari_
not modelled
5.3
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)65 d1mg7a1
not modelled
5.2
33
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Early switch protein XOL-1, N-terminal domain66 c3q13A_
not modelled
5.1
21
PDB header: cell adhesionChain: A: PDB Molecule: spondin-1;PDBTitle: the structure of the ca2+-binding, glycosylated f-spondin domain of f-2 spondin, a c2-domain variant from extracellular matrix
67 c3oqlA_
not modelled
5.1
27
PDB header: transcriptionChain: A: PDB Molecule: tena homolog;PDBTitle: crystal structure of a tena homolog (pspto1738) from pseudomonas2 syringae pv. tomato str. dc3000 at 2.54 a resolution
68 d1buna_
not modelled
5.1
17
Fold: Phospholipase A2, PLA2Superfamily: Phospholipase A2, PLA2Family: Vertebrate phospholipase A2