1 c2pmzN_
33.6
31
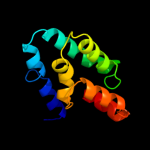
PDB header: translation, transferaseChain: N: PDB Molecule: dna-directed rna polymerase subunit n;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
2 d1ef4a_
26.8
33
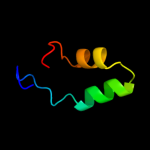
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB103 c3f6wE_
26.3
13
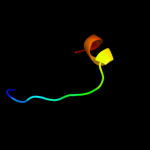
PDB header: dna binding proteinChain: E: PDB Molecule: xre-family like protein;PDBTitle: xre-family like protein from pseudomonas syringae pv. tomato str.2 dc3000
4 d2qkwa1
21.4
17
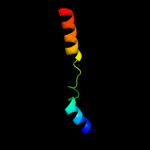
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Avirulence protein AvrPtoFamily: Avirulence protein AvrPto5 c2qkwA_
21.4
17
PDB header: transferaseChain: A: PDB Molecule: avirulence protein;PDBTitle: structural basis for activation of plant immunity by2 bacterial effector protein avrpto
6 d1qi9a_
17.1
23
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)7 d1gw5b_
17.1
16
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: Clathrin adaptor core protein8 c2zopA_
15.8
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative uncharacterized protein tthb164;PDBTitle: x-ray crystal structure of a crispr-associated cmr5 family2 protein from thermus thermophilus hb8
9 d1twfj_
15.7
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: RNA polymerase subunit RPB10Family: RNA polymerase subunit RPB1010 d2bosa_
13.5
47
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits11 c1m5iA_
13.4
24
PDB header: antitumor proteinChain: A: PDB Molecule: apc protein;PDBTitle: crystal structure of the coiled coil region 129-250 of the2 tumor suppressor gene product apc
12 c3ol4B_
13.1
21
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
13 c3omtA_
13.0
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: putative antitoxin component, chu_2935 protein, from xre family from2 prevotella buccae.
14 d1qhba_
12.7
21
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)15 c3k2hA_
11.9
28
PDB header: transferaseChain: A: PDB Molecule: dihydrofolate reductase/thymidylate synthase;PDBTitle: co-crystal structure of dihydrofolate reductase/thymidylate synthase2 from babesia bovis with dump, pemetrexed and nadp
16 d1utxa_
11.7
23
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: SinR domain-like17 d1up8a_
11.7
19
Fold: Acid phosphatase/Vanadium-dependent haloperoxidaseSuperfamily: Acid phosphatase/Vanadium-dependent haloperoxidaseFamily: Haloperoxidase (bromoperoxidase)18 c3fm8A_
11.2
60
PDB header: transport protein/hydrolase activatorChain: A: PDB Molecule: kinesin-like protein kif13b;PDBTitle: crystal structure of full length centaurin alpha-1 bound with the fha2 domain of kif13b (capri target)
19 c3jsuA_
11.0
28
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: dihydrofolate reductase-thymidylate synthase;PDBTitle: quadruple mutant(n51i+c59r+s108n+i164l) plasmodium falciparum2 dihydrofolate reductase-thymidylate synthase(pfdhfr-ts) complexed3 with qn254, nadph, and dump
20 c2dunA_
10.7
16
PDB header: transferaseChain: A: PDB Molecule: dna polymerase mu;PDBTitle: solution structure of brct domain of dna polymerase mu
21 d2coba1
not modelled
10.0
9
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Psq domain22 d1biaa1
not modelled
9.7
28
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Biotin repressor-like23 c2of3A_
not modelled
9.4
10
PDB header: structural protein, cell cycleChain: A: PDB Molecule: zyg-9;PDBTitle: tog domain structure from c.elegans zyg9
24 d1r4pb_
not modelled
9.2
47
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits25 c1yn3A_
not modelled
9.1
19
PDB header: toxin, protein bindingChain: A: PDB Molecule: truncated cell surface protein map-w;PDBTitle: crystal structures of eap domains from staphylococcus2 aureus reveal an unexpected homology to bacterial3 superantigens
26 c2kygC_
not modelled
9.0
19
PDB header: protein bindingChain: C: PDB Molecule: protein cbfa2t1;PDBTitle: structure of the aml1-eto nervy domain - pka(riia) complex and its2 contribution to aml1-eto activity
27 d2dk5a1
not modelled
8.7
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RPO3F domain-like28 d1mhyd_
not modelled
8.6
14
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like29 d1ktba2
not modelled
8.5
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: Amylase, catalytic domain30 c1yn5B_
not modelled
8.2
28
PDB header: unknown functionChain: B: PDB Molecule: eaph2;PDBTitle: crystal structures of eap domains from staphylococcus2 aureus reveal an unexpected homology to bacterial3 superantigens
31 c2jspA_
not modelled
8.2
5
PDB header: gene regulationChain: A: PDB Molecule: transcriptional regulatory protein ros;PDBTitle: the prokaryotic cys2his2 zinc finger adopts a novel fold as2 revealed by the nmr structure of a. tumefaciens ros dna3 binding domain
32 c1yn4A_
not modelled
8.2
20
PDB header: unknown functionChain: A: PDB Molecule: eaph1;PDBTitle: crystal structures of eap domains from staphylococcus2 aureus reveal an unexpected homology to bacterial3 superantigens
33 c2yu3A_
not modelled
7.5
15
PDB header: transcriptionChain: A: PDB Molecule: dna-directed rna polymerase iii 39 kdaPDBTitle: solution structure of the domain swapped wingedhelix in dna-2 directed rna polymerase iii 39 kda polypeptide
34 c3clbA_
not modelled
7.3
34
PDB header: oxidoreductase, transferaseChain: A: PDB Molecule: dhfr-ts;PDBTitle: structure of bifunctional tcdhfr-ts in complex with tmq
35 c2ewtA_
not modelled
7.1
16
PDB header: dna binding proteinChain: A: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of the dna-binding domain of bldd
36 c3e7lD_
not modelled
6.6
20
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
37 c3mklB_
not modelled
6.6
14
PDB header: transcription regulatorChain: B: PDB Molecule: hth-type transcriptional regulator gadx;PDBTitle: crystal structure of dna-binding transcriptional dual regulator from2 escherichia coli k-12
38 c2yujA_
not modelled
6.6
19
PDB header: protein bindingChain: A: PDB Molecule: ubiquitin fusion degradation 1-like;PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
39 c2k1gA_
not modelled
6.6
13
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein spr;PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
40 c3q9dB_
not modelled
6.3
32
PDB header: unknown functionChain: B: PDB Molecule: protein cpn_0803/cp_1068/cpj0803/cpb0832;PDBTitle: crystal structure of cpn0803 from c. pneumoniae.
41 d1c4qa_
not modelled
6.1
33
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits42 d2ixna1
not modelled
5.7
22
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like43 d2huha1
not modelled
5.5
38
Fold: C2 domain-likeSuperfamily: Smr-associated domain-likeFamily: Smr-associated domain44 d2nrac2
not modelled
5.4
35
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Replication initiation protein45 d1tkna_
not modelled
5.4
19
Fold: STAT-likeSuperfamily: CAPPD, an extracellular domain of amyloid beta A4 proteinFamily: CAPPD, an extracellular domain of amyloid beta A4 protein46 c2nogA_
not modelled
5.4
12
PDB header: dna binding proteinChain: A: PDB Molecule: iswi protein;PDBTitle: sant domain structure of xenopus remodeling factor iswi
47 c1ktbA_
not modelled
5.2
12
PDB header: hydrolaseChain: A: PDB Molecule: alpha-n-acetylgalactosaminidase;PDBTitle: the structure of alpha-n-acetylgalactosaminidase
48 c1w9zB_
not modelled
5.2
45
PDB header: virus coat proteinChain: B: PDB Molecule: vp9;PDBTitle: structure of bannavirus vp9
49 c2zb9A_
not modelled
5.2
15
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of tetr family transcription regulator2 sco0332
50 c2qw6A_
not modelled
5.2
26
PDB header: hydrolaseChain: A: PDB Molecule: aaa atpase, central region;PDBTitle: crystal structure of the c-terminal domain of an aaa atpase from2 enterococcus faecium do
51 d2qw6a1
not modelled
5.2
26
Fold: post-AAA+ oligomerization domain-likeSuperfamily: post-AAA+ oligomerization domain-likeFamily: MgsA/YrvN C-terminal domain-like52 d1sr9a3
not modelled
5.1
13
Fold: 2-isopropylmalate synthase LeuA, allosteric (dimerisation) domainSuperfamily: 2-isopropylmalate synthase LeuA, allosteric (dimerisation) domainFamily: 2-isopropylmalate synthase LeuA, allosteric (dimerisation) domain