| 1 |
|
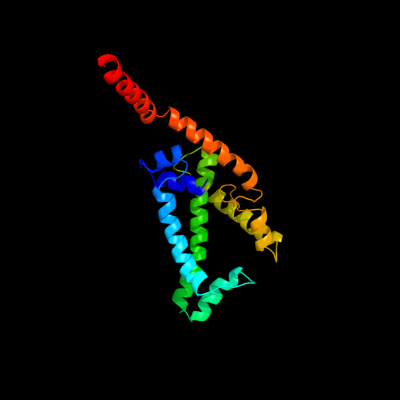
PDB 2w8a chain C
Region: 31 - 228
Aligned: 181
Modelled: 197
Confidence: 16.2%
Identity: 10%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 2 |
|
PDB 3hd6 chain A
Region: 108 - 228
Aligned: 118
Modelled: 121
Confidence: 11.6%
Identity: 12%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 3 |
|
PDB 2b2h chain A
Region: 82 - 228
Aligned: 147
Modelled: 147
Confidence: 9.6%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter;
PDBTitle: ammonium transporter amt-1 from a. fulgidus (as)
Phyre2
| 4 |
|
PDB 3b9y chain A
Region: 108 - 240
Aligned: 124
Modelled: 133
Confidence: 8.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 72 - 250
Aligned: 179
Modelled: 179
Confidence: 8.1%
Identity: 8%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 79 - 302
Aligned: 199
Modelled: 207
Confidence: 7.4%
Identity: 14%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 1xme chain A domain 1
Region: 177 - 261
Aligned: 85
Modelled: 85
Confidence: 7.0%
Identity: 6%
Fold: Cytochrome c oxidase subunit I-like
Superfamily: Cytochrome c oxidase subunit I-like
Family: Cytochrome c oxidase subunit I-like
Phyre2
| 8 |
|
PDB 3eh4 chain A
Region: 177 - 261
Aligned: 85
Modelled: 85
Confidence: 6.3%
Identity: 6%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c oxidase subunit 1;
PDBTitle: structure of the reduced form of cytochrome ba3 oxidase from thermus2 thermophilus
Phyre2
| 9 |
|
PDB 2vof chain A
Region: 62 - 99
Aligned: 38
Modelled: 37
Confidence: 6.0%
Identity: 11%
PDB header:apoptosis
Chain: A: PDB Molecule:bcl-2-related protein a1;
PDBTitle: structure of mouse a1 bound to the puma bh3-domain
Phyre2
| 10 |
|
PDB 2knc chain A
Region: 180 - 222
Aligned: 43
Modelled: 43
Confidence: 5.8%
Identity: 7%
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
Phyre2
| 11 |
|
PDB 2oar chain A
Region: 180 - 236
Aligned: 57
Modelled: 57
Confidence: 5.8%
Identity: 12%
PDB header:membrane protein
Chain: A: PDB Molecule:large-conductance mechanosensitive channel;
PDBTitle: mechanosensitive channel of large conductance (mscl)
Phyre2