| 1 |
|
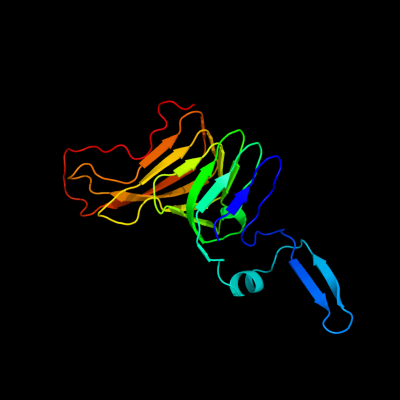
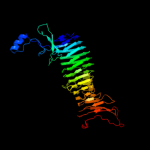
PDB 3h09 chain B
Region: 230 - 411
Aligned: 180
Modelled: 182
Confidence: 99.2%
Identity: 17%
PDB header:hydrolase
Chain: B: PDB Molecule:immunoglobulin a1 protease;
PDBTitle: the structure of haemophilus influenzae iga1 protease
Phyre2
| 2 |
|
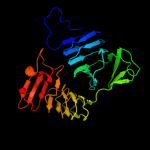
PDB 1dab chain A
Region: 44 - 412
Aligned: 365
Modelled: 365
Confidence: 98.9%
Identity: 11%
Fold: Single-stranded right-handed beta-helix
Superfamily: Pectin lyase-like
Family: Virulence factor P.69 pertactin
Phyre2
| 3 |
|
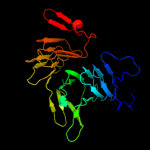
PDB 3ml3 chain A
Region: 309 - 412
Aligned: 104
Modelled: 104
Confidence: 98.6%
Identity: 16%
PDB header:protein transport
Chain: A: PDB Molecule:outer membrane protein icsa autotransporter;
PDBTitle: crystal structure of the icsa autochaperone region
Phyre2
| 4 |
|
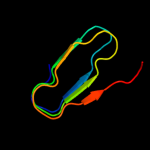
PDB 3syj chain A
Region: 145 - 411
Aligned: 266
Modelled: 267
Confidence: 98.2%
Identity: 9%
PDB header:cell adhesion
Chain: A: PDB Molecule:adhesion and penetration protein autotransporter;
PDBTitle: crystal structure of the haemophilus influenzae hap adhesin
Phyre2
| 5 |
|
PDB 3ak5 chain B
Region: 10 - 415
Aligned: 403
Modelled: 404
Confidence: 89.9%
Identity: 14%
PDB header:hydrolase
Chain: B: PDB Molecule:hemoglobin-binding protease hbp;
PDBTitle: hemoglobin protease (hbp) passenger missing domain-2
Phyre2
| 6 |
|
PDB 2zj6 chain A
Region: 2 - 259
Aligned: 258
Modelled: 258
Confidence: 57.8%
Identity: 12%
PDB header:hydrolase
Chain: A: PDB Molecule:lipase;
PDBTitle: crystal structure of d337a mutant of pseudomonas sp. mis38 lipase
Phyre2
| 7 |
|
PDB 2qub chain G
Region: 1 - 245
Aligned: 243
Modelled: 245
Confidence: 28.5%
Identity: 9%
PDB header:hydrolase
Chain: G: PDB Molecule:extracellular lipase;
PDBTitle: crystal structure of extracellular lipase lipa from serratia2 marcescens
Phyre2
| 8 |
|
PDB 1kap chain P domain 1
Region: 1 - 50
Aligned: 50
Modelled: 50
Confidence: 18.5%
Identity: 20%
Fold: Single-stranded right-handed beta-helix
Superfamily: beta-Roll
Family: Serralysin-like metalloprotease, C-terminal domain
Phyre2
| 9 |
|
PDB 2agm chain A
Region: 1 - 65
Aligned: 65
Modelled: 65
Confidence: 16.2%
Identity: 15%
PDB header:isomerase
Chain: A: PDB Molecule:poly(beta-d-mannuronate) c5 epimerase 4;
PDBTitle: solution structure of the r-module from alge4
Phyre2