| 1 |
|
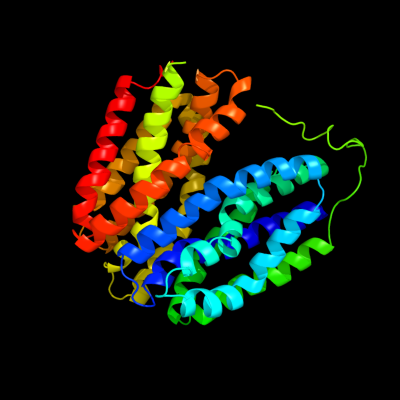
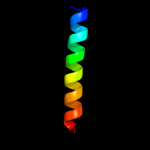
PDB 1pv7 chain A
Region: 11 - 397
Aligned: 387
Modelled: 387
Confidence: 100.0%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 2 |
|
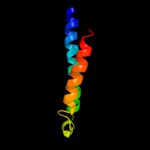
PDB 1pw4 chain A
Region: 2 - 396
Aligned: 393
Modelled: 395
Confidence: 100.0%
Identity: 10%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 3 |
|
PDB 2gfp chain A
Region: 16 - 385
Aligned: 368
Modelled: 370
Confidence: 100.0%
Identity: 9%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 4 |
|
PDB 3o7p chain A
Region: 14 - 395
Aligned: 379
Modelled: 379
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 18 - 392
Aligned: 374
Modelled: 375
Confidence: 99.9%
Identity: 11%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 2w8a chain C
Region: 17 - 214
Aligned: 198
Modelled: 198
Confidence: 11.8%
Identity: 13%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 7 |
|
PDB 2g9p chain A
Region: 68 - 81
Aligned: 14
Modelled: 14
Confidence: 11.8%
Identity: 14%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 8 |
|
PDB 2xq2 chain A
Region: 47 - 210
Aligned: 162
Modelled: 164
Confidence: 8.7%
Identity: 7%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 9 |
|
PDB 2ks1 chain B
Region: 373 - 397
Aligned: 25
Modelled: 25
Confidence: 8.4%
Identity: 20%
PDB header:transferase
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
Phyre2
| 10 |
|
PDB 2bbj chain B
Region: 337 - 396
Aligned: 58
Modelled: 60
Confidence: 8.0%
Identity: 9%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 11 |
|
PDB 2vut chain I domain 1
Region: 6 - 15
Aligned: 10
Modelled: 10
Confidence: 7.2%
Identity: 30%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: Erythroid transcription factor GATA-1
Phyre2
| 12 |
|
PDB 1y0j chain A domain 1
Region: 6 - 15
Aligned: 10
Modelled: 10
Confidence: 5.9%
Identity: 40%
Fold: Glucocorticoid receptor-like (DNA-binding domain)
Superfamily: Glucocorticoid receptor-like (DNA-binding domain)
Family: Erythroid transcription factor GATA-1
Phyre2