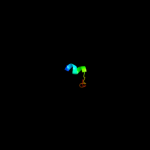| 1 |
|
PDB 3bdr chain A
Region: 12 - 21
Aligned: 10
Modelled: 10
Confidence: 37.0%
Identity: 60%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:ycf58 protein;
PDBTitle: crystal structure of fatty acid-binding protein-like ycf582 from thermosynecoccus elongatus. northeast structural3 genomics consortium target ter13.
Phyre2
| 2 |
|
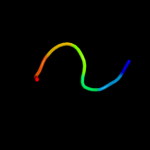
PDB 2y69 chain X
Region: 9 - 27
Aligned: 19
Modelled: 19
Confidence: 15.1%
Identity: 32%
PDB header:electron transport
Chain: X: PDB Molecule:cytochrome c oxidase polypeptide 7b;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
Phyre2
| 3 |
|
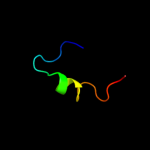
PDB 1v54 chain K
Region: 9 - 27
Aligned: 19
Modelled: 19
Confidence: 15.1%
Identity: 32%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIb
Family: Mitochondrial cytochrome c oxidase subunit VIIb
Phyre2
| 4 |
|
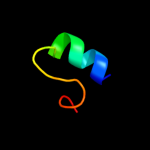
PDB 2yum chain A
Region: 5 - 27
Aligned: 23
Modelled: 23
Confidence: 10.8%
Identity: 17%
PDB header:transcription
Chain: A: PDB Molecule:zinc finger zz-type-containing protein 3;
PDBTitle: solution structure of the myb-like dna-binding domain of2 human zzz3 protein
Phyre2
| 5 |
|
PDB 1zec chain A
Region: 19 - 25
Aligned: 7
Modelled: 7
Confidence: 7.8%
Identity: 71%
PDB header:viral peptide
Chain: A: PDB Molecule:nef1-25;
PDBTitle: nmr solution structure of nef1-25, 20 structures
Phyre2
| 6 |
|
PDB 1h6v chain A domain 3
Region: 16 - 26
Aligned: 11
Modelled: 11
Confidence: 7.7%
Identity: 36%
Fold: CO dehydrogenase flavoprotein C-domain-like
Superfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Family: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
Phyre2
| 7 |
|
PDB 1x41 chain A domain 1
Region: 11 - 27
Aligned: 17
Modelled: 17
Confidence: 7.4%
Identity: 29%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Homeodomain
Phyre2
| 8 |
|
PDB 1fdf chain A
Region: 12 - 22
Aligned: 11
Modelled: 11
Confidence: 6.7%
Identity: 55%
PDB header:signaling protein
Chain: A: PDB Molecule:rhodopsin;
PDBTitle: helix 7 bovine rhodopsin
Phyre2
| 9 |
|
PDB 1uw4 chain A
Region: 1 - 23
Aligned: 22
Modelled: 23
Confidence: 6.4%
Identity: 14%
Fold: Ferredoxin-like
Superfamily: RNA-binding domain, RBD
Family: Smg-4/UPF3
Phyre2
| 10 |
|
PDB 1wgx chain A
Region: 10 - 27
Aligned: 18
Modelled: 18
Confidence: 6.1%
Identity: 28%
Fold: DNA/RNA-binding 3-helical bundle
Superfamily: Homeodomain-like
Family: Myb/SANT domain
Phyre2
| 11 |
|
PDB 2wj8 chain N
Region: 8 - 18
Aligned: 11
Modelled: 11
Confidence: 6.0%
Identity: 64%
PDB header:rna binding protein/rna
Chain: N: PDB Molecule:nucleoprotein;
PDBTitle: respiratory syncitial virus ribonucleoprotein
Phyre2