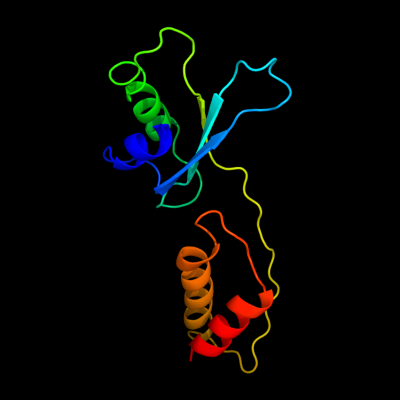
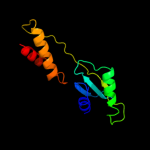
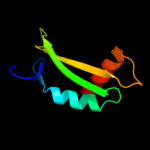
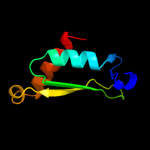
1 c3h3yF_
98.1
13
PDB header: viral proteinChain: F: PDB Molecule: baseplate structural protein gp6;PDBTitle: fitting of the gp6 crystal structure into 3d cryo-em2 reconstruction of bacteriophage t4 star-shaped baseplate
2 c3etcB_
71.0
7
PDB header: ligaseChain: B: PDB Molecule: amp-binding protein;PDBTitle: 2.1 a structure of acyl-adenylate synthetase from methanosarcina2 acetivorans containing a link between lys256 and cys298
3 c3eynB_
45.1
9
PDB header: ligaseChain: B: PDB Molecule: acyl-coenzyme a synthetase acsm2a;PDBTitle: crystal structure of human acyl-coa synthetase medium-chain2 family member 2a (l64p mutation) in a complex with coa
4 c3iplB_
44.2
15
PDB header: ligaseChain: B: PDB Molecule: 2-succinylbenzoate--coa ligase;PDBTitle: crystal structure of o-succinylbenzoic acid-coa ligase from2 staphylococcus aureus subsp. aureus mu50
5 c3nyrA_
39.7
11
PDB header: ligaseChain: A: PDB Molecule: malonyl-coa ligase;PDBTitle: malonyl-coa ligase ternary product complex with malonyl-coa and amp2 bound
6 c3dhvA_
33.7
5
PDB header: ligaseChain: A: PDB Molecule: d-alanine-poly(phosphoribitol) ligase;PDBTitle: crystal structure of dlta protein in complex with d-alanine2 adenylate
7 c2d1tA_
32.6
9
PDB header: oxidoreductaseChain: A: PDB Molecule: luciferin 4-monooxygenase;PDBTitle: crystal structure of the thermostable japanese firefly2 luciferase red-color emission s286n mutant complexed with3 high-energy intermediate analogue
8 d1amua_
32.0
8
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like9 d1lcia_
25.8
9
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like10 c1amuB_
25.6
8
PDB header: peptide synthetaseChain: B: PDB Molecule: gramicidin synthetase 1;PDBTitle: phenylalanine activating domain of gramicidin synthetase 12 in a complex with amp and phenylalanine
11 c3e7wA_
23.2
9
PDB header: ligaseChain: A: PDB Molecule: d-alanine--poly(phosphoribitol) ligase subunit 1;PDBTitle: crystal structure of dlta: implications for the reaction2 mechanism of non-ribosomal peptide synthetase (nrps)3 adenylation domains
12 d1mdba_
23.2
7
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like13 c3gqwB_
21.4
12
PDB header: ligaseChain: B: PDB Molecule: fatty acid amp ligase;PDBTitle: crystal structure of a fatty acid amp ligase from e. coli with an acyl2 adenylate product bound
14 d1u9la_
20.9
23
Fold: SAM domain-likeSuperfamily: Rad51 N-terminal domain-likeFamily: NusA extra C-terminal domains15 d1pg4a_
18.8
8
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like16 d1xhja_
17.6
18
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like17 c3l8cA_
12.5
13
PDB header: ligaseChain: A: PDB Molecule: d-alanine--poly(phosphoribitol) ligase subunit 1;PDBTitle: structure of probable d-alanine--poly(phosphoribitol) ligase2 subunit-1 from streptococcus pyogenes
18 c2aj1A_
10.0
6
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
19 d2qifa1
9.8
3
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain20 c2l3mA_
8.9
10
PDB header: metal binding proteinChain: A: PDB Molecule: copper-ion-binding protein;PDBTitle: solution structure of the putative copper-ion-binding protein from2 bacillus anthracis str. ames
21 c3ni2A_
not modelled
8.8
7
PDB header: ligaseChain: A: PDB Molecule: 4-coumarate:coa ligase;PDBTitle: crystal structures and enzymatic mechanisms of a populus tomentosa 4-2 coumarate:coa ligase
22 d1veha_
not modelled
8.6
12
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Fe-S cluster assembly (FSCA) domain-likeFamily: NifU C-terminal domain-like23 d1p6ta2
not modelled
7.3
13
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain24 c3qovD_
not modelled
7.2
9
PDB header: ligaseChain: D: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of a hypothetical acyl-coa ligase (bt_0428) from2 bacteroides thetaiotaomicron vpi-5482 at 2.20 a resolution
25 d1osda_
not modelled
7.0
13
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain26 c2y27B_
not modelled
7.0
6
PDB header: ligaseChain: B: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: crystal structure of paak1 in complex with atp from2 burkholderia cenocepacia
27 d1sb6a_
not modelled
7.0
10
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain28 c1y96D_
not modelled
6.8
9
PDB header: rna binding proteinChain: D: PDB Molecule: gem-associated protein 7;PDBTitle: crystal structure of the gemin6/gemin7 heterodimer from the2 human smn complex
29 d1ry2a_
not modelled
6.7
14
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like30 c3l48B_
not modelled
6.6
16
PDB header: transport proteinChain: B: PDB Molecule: outer membrane usher protein papc;PDBTitle: crystal structure of the c-terminal domain of the papc usher
31 c2k2pA_
not modelled
6.5
10
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu1203;PDBTitle: solution nmr structure of protein atu1203 from agrobacterium2 tumefaciens. northeast structural genomics consortium (nesg) target3 att10, ontario center for structural proteomics target atc1183
32 c2kt2A_
not modelled
6.2
19
PDB header: oxidoreductaseChain: A: PDB Molecule: mercuric reductase;PDBTitle: structure of nmera, the n-terminal hma domain of tn501 mercuric2 reductase
33 c2ofhX_
not modelled
6.1
6
PDB header: hydrolase, membrane proteinChain: X: PDB Molecule: zinc-transporting atpase;PDBTitle: solution structure of the n-terminal domain of the zinc(ii) atpase2 ziaa in its apo form
34 c2v7bB_
not modelled
6.1
7
PDB header: ligaseChain: B: PDB Molecule: benzoate-coenzyme a ligase;PDBTitle: crystal structures of a benzoate coa ligase from2 burkholderia xenovorans lb400
35 d1mnta_
not modelled
6.1
6
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: Arc/Mnt-like phage repressors36 c1yjrA_
not modelled
5.7
10
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the apo form of the sixth soluble2 domain a69p mutant of menkes protein
37 c3dxsX_
not modelled
5.6
10
PDB header: hydrolaseChain: X: PDB Molecule: copper-transporting atpase ran1;PDBTitle: crystal structure of a copper binding domain from hma7, a p-2 type atpase
38 d1cpza_
not modelled
5.3
19
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain39 c2kkhA_
not modelled
5.2
10
PDB header: metal transportChain: A: PDB Molecule: putative heavy metal transporter;PDBTitle: structure of the zinc binding domain of the atpase hma4
40 c2ga7A_
not modelled
5.2
16
PDB header: hydrolaseChain: A: PDB Molecule: copper-transporting atpase 1;PDBTitle: solution structure of the copper(i) form of the third metal-2 binding domain of atp7a protein (menkes disease protein)
41 c3laxA_
not modelled
5.2
3
PDB header: ligaseChain: A: PDB Molecule: phenylacetate-coenzyme a ligase;PDBTitle: the crystal structure of a domain of phenylacetate-coenzyme2 a ligase from bacteroides vulgatus atcc 8482
42 c3qu3A_
not modelled
5.1
9
PDB header: dna binding proteinChain: A: PDB Molecule: interferon regulatory factor 7;PDBTitle: crystal structure of irf-7 dbd apo form
43 c2uwjE_
not modelled
5.0
12
PDB header: chaperoneChain: E: PDB Molecule: type iii export protein psce;PDBTitle: structure of the heterotrimeric complex which regulates2 type iii secretion needle formation
44 d1q8la_
not modelled
5.0
16
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain