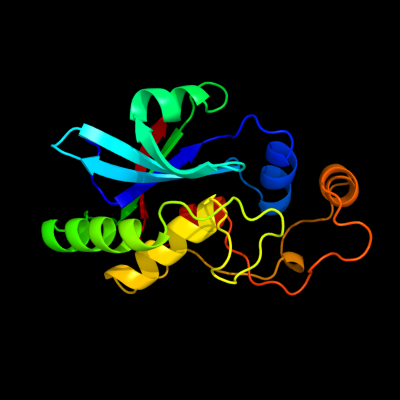
| 1 | d2if6a1
|
|
 |
100.0 |
25 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:YiiX-like |
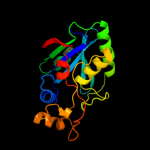
| 2 | c3kw0D_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:cysteine peptidase;
PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
|
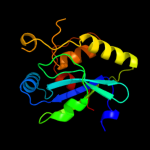
| 3 | c2p1gA_
|
|
 |
98.1 |
23 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative xylanase;
PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
|
| 4 | c3h41A_
|
|
 |
97.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:nlp/p60 family protein;
PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
|
| 5 | c2k1gA_
|
|
 |
97.5 |
22 |
PDB header:lipoprotein
Chain: A: PDB Molecule:lipoprotein spr;
PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
|
| 6 | c2kytA_
|
|
 |
97.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:group xvi phospholipase a2;
PDBTitle: solution struture of the h-rev107 n-terminal domain
|
| 7 | c2fg0B_
|
|
 |
97.3 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:cog0791: cell wall-associated hydrolases (invasion-
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
|
| 8 | d2evra2
|
|
 |
97.2 |
26 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:NlpC/P60 |
| 9 | c2im9A_
|
|
 |
97.0 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of protein lpg0564 from legionella pneumophila str.2 philadelphia 1, pfam duf1460
|
| 10 | d2im9a1
|
|
 |
97.0 |
13 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:Lpg0564-like |
| 11 | c3gt2A_
|
|
 |
97.0 |
10 |
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
|
| 12 | c2xivA_
|
|
 |
96.7 |
18 |
PDB header:structural protein
Chain: A: PDB Molecule:hypothetical invasion protein;
PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
|
| 13 | c3i86A_
|
|
 |
96.7 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
|
| 14 | c3pbiA_
|
|
 |
96.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:invasion protein;
PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
|
| 15 | c3npfB_
|
|
 |
96.1 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative dipeptidyl-peptidase vi;
PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
|
| 16 | d2io8a2
|
|
 |
93.0 |
25 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:CHAP domain |
| 17 | c3m1uB_
|
|
 |
91.0 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative gamma-d-glutamyl-l-diamino acid endopeptidase;
PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
|
| 18 | c2vpmB_
|
|
 |
83.5 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:trypanothione synthetase;
PDBTitle: trypanothione synthetase
|
| 19 | c2ioaA_
|
|
 |
83.1 |
26 |
PDB header:ligase, hydrolase
Chain: A: PDB Molecule:bifunctional glutathionylspermidine
PDBTitle: e. coli bifunctional glutathionylspermidine2 synthetase/amidase incomplex with mg2+ and adp and3 phosphinate inhibitor
|
| 20 | c2k3aA_
|
|
 |
66.9 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:chap domain protein;
PDBTitle: nmr solution structure of staphylococcus saprophyticus chap2 (cysteine, histidine-dependent amidohydrolases/peptidases)3 domain protein. northeast structural genomics consortium4 target syr11
|
| 21 | d1wh7a_ |
|
not modelled |
44.2 |
24 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:Homeodomain |
| 22 | d1igna2 |
|
not modelled |
41.0 |
9 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Homeodomain-like
Family:DNA-binding domain of rap1 |
| 23 | d1wfza_ |
|
not modelled |
29.0 |
35 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 24 | c3nnlB_ |
|
not modelled |
27.2 |
33 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:cura;
PDBTitle: halogenase domain from cura module (crystal form iii)
|
| 25 | c2z7eB_ |
|
not modelled |
25.4 |
12 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:nifu-like protein;
PDBTitle: crystal structure of aquifex aeolicus iscu with bound [2fe-2 2s] cluster
|
| 26 | d2glza1 |
|
not modelled |
22.5 |
10 |
Fold:FwdE/GAPDH domain-like
Superfamily:FwdE-like
Family:FwdE-like |
| 27 | d2gy9q1 |
|
not modelled |
19.3 |
30 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 28 | d1hkva1 |
|
not modelled |
18.0 |
20 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 29 | d1wpna_ |
|
not modelled |
17.1 |
15 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 30 | d1vbga2 |
|
not modelled |
15.4 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 31 | c1khiA_ |
|
not modelled |
15.2 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:hex1;
PDBTitle: crystal structure of hex1
|
| 32 | d1h2ka_ |
|
not modelled |
14.7 |
24 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Hypoxia-inducible factor HIF ihhibitor (FIH1) |
| 33 | d2z1ea2 |
|
not modelled |
14.7 |
24 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 34 | c2zkqq_ |
|
not modelled |
14.2 |
30 |
PDB header:ribosomal protein/rna
Chain: Q: PDB Molecule:
PDBTitle: structure of a mammalian ribosomal 40s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
|
| 35 | d1khia1 |
|
not modelled |
13.5 |
15 |
Fold:SH3-like barrel
Superfamily:Translation proteins SH3-like domain
Family:eIF5a N-terminal domain-like |
| 36 | d1zyma2 |
|
not modelled |
13.3 |
14 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system |
| 37 | c3bbnQ_ |
|
not modelled |
13.3 |
15 |
PDB header:ribosome
Chain: Q: PDB Molecule:ribosomal protein s17;
PDBTitle: homology model for the spinach chloroplast 30s subunit2 fitted to 9.4a cryo-em map of the 70s chlororibosome.
|
| 38 | c3zvmA_ |
|
not modelled |
13.0 |
19 |
PDB header:hydrolase/transferase/dna
Chain: A: PDB Molecule:bifunctional polynucleotide phosphatase/kinase;
PDBTitle: the structural basis for substrate recognition by mammalian2 polynucleotide kinase 3' phosphatase
|
| 39 | d1r9pa_ |
|
not modelled |
12.7 |
29 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 40 | c2obnA_ |
|
not modelled |
12.6 |
19 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a duf1611 family protein (ava_3511) from anabaena2 variabilis atcc 29413 at 2.30 a resolution
|
| 41 | d1k20a_ |
|
not modelled |
12.4 |
26 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 42 | d1asua_ |
|
not modelled |
12.4 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Ribonuclease H-like
Family:Retroviral integrase, catalytic domain |
| 43 | c3k1rA_ |
|
not modelled |
12.1 |
19 |
PDB header:structural protein
Chain: A: PDB Molecule:harmonin;
PDBTitle: structure of harmonin npdz1 in complex with the sam-pbm of2 sans
|
| 44 | d1kbla2 |
|
not modelled |
11.8 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 45 | c1s1hQ_ |
|
not modelled |
11.6 |
20 |
PDB header:ribosome
Chain: Q: PDB Molecule:40s ribosomal protein s11;
PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
|
| 46 | c3k2oB_ |
|
not modelled |
11.6 |
41 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:bifunctional arginine demethylase and lysyl-hydroxylase
PDBTitle: structure of an oxygenase
|
| 47 | d1h6za2 |
|
not modelled |
11.6 |
13 |
Fold:The "swivelling" beta/beta/alpha domain
Superfamily:Phosphohistidine domain
Family:Pyruvate phosphate dikinase, central domain |
| 48 | c2x35A_ |
|
not modelled |
11.5 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:peregrin;
PDBTitle: molecular basis of histone h3k36me3 recognition by the pwwp2 domain of brpf1.
|
| 49 | c2zpmA_ |
|
not modelled |
11.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:regulator of sigma e protease;
PDBTitle: crystal structure analysis of pdz domain b
|
| 50 | d1xjsa_ |
|
not modelled |
11.3 |
12 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 51 | c2opwA_ |
|
not modelled |
11.0 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:phyhd1 protein;
PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
|
| 52 | d2qamc2 |
|
not modelled |
10.8 |
28 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 53 | d1rl2a2 |
|
not modelled |
10.7 |
17 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 54 | c1zx6A_ |
|
not modelled |
10.5 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:ypr154wp;
PDBTitle: high-resolution crystal structure of yeast pin3 sh3 domain
|
| 55 | d2uubq1 |
|
not modelled |
10.3 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 56 | d1vcta2 |
|
not modelled |
10.2 |
8 |
Fold:TrkA C-terminal domain-like
Superfamily:TrkA C-terminal domain-like
Family:TrkA C-terminal domain-like |
| 57 | d2hawa1 |
|
not modelled |
9.8 |
15 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 58 | d1su0b_ |
|
not modelled |
9.5 |
35 |
Fold:SufE/NifU
Superfamily:SufE/NifU
Family:NifU/IscU domain |
| 59 | d1i94q_ |
|
not modelled |
9.3 |
20 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 60 | c1ezaA_ |
|
not modelled |
9.0 |
12 |
PDB header:phosphotransferase
Chain: A: PDB Molecule:enzyme i;
PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
|
| 61 | d2f4pa1 |
|
not modelled |
9.0 |
18 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:TM1287-like |
| 62 | d1ripa_ |
|
not modelled |
8.8 |
30 |
Fold:OB-fold
Superfamily:Nucleic acid-binding proteins
Family:Cold shock DNA-binding domain-like |
| 63 | c2wkdA_ |
|
not modelled |
8.7 |
27 |
PDB header:dna binding protein
Chain: A: PDB Molecule:orf34p2;
PDBTitle: crystal structure of a double ile-to-met mutant of protein2 orf34 from lactococcus phage p2
|
| 64 | d2fcta1 |
|
not modelled |
8.7 |
13 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:PhyH-like |
| 65 | d2hg6a1 |
|
not modelled |
8.6 |
50 |
Fold:PA1123-like
Superfamily:PA1123-like
Family:PA1123-like |
| 66 | d1i74a_ |
|
not modelled |
8.3 |
28 |
Fold:DHH phosphoesterases
Superfamily:DHH phosphoesterases
Family:Manganese-dependent inorganic pyrophosphatase (family II) |
| 67 | c3mt1B_ |
|
not modelled |
8.2 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:putative carboxynorspermidine decarboxylase protein;
PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
|
| 68 | d1ntha_ |
|
not modelled |
8.1 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Monomethylamine methyltransferase MtmB
Family:Monomethylamine methyltransferase MtmB |
| 69 | d1vrba1 |
|
not modelled |
8.0 |
6 |
Fold:Double-stranded beta-helix
Superfamily:Clavaminate synthase-like
Family:Asparaginyl hydroxylase-like |
| 70 | d2nlua1 |
|
not modelled |
8.0 |
38 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:PWWP domain |
| 71 | d2zoda2 |
|
not modelled |
8.0 |
6 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 72 | d1hyoa2 |
|
not modelled |
7.6 |
17 |
Fold:FAH
Superfamily:FAH
Family:FAH |
| 73 | d1wpga1 |
|
not modelled |
7.4 |
28 |
Fold:Double-stranded beta-helix
Superfamily:Calcium ATPase, transduction domain A
Family:Calcium ATPase, transduction domain A |
| 74 | c3al6A_ |
|
not modelled |
7.3 |
41 |
PDB header:unknown function
Chain: A: PDB Molecule:jmjc domain-containing protein c2orf60;
PDBTitle: crystal structure of human tyw5
|
| 75 | c1nhgD_ |
|
not modelled |
7.2 |
9 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-acyl carrier reductase;
PDBTitle: crystal structure analysis of plasmodium falciparum enoyl-2 acyl-carrier-protein reductase with triclosan
|
| 76 | d1fc6a3 |
|
not modelled |
7.2 |
30 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:Tail specific protease PDZ domain |
| 77 | c1yj5B_ |
|
not modelled |
7.1 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:5' polynucleotide kinase-3' phosphatase catalytic domain;
PDBTitle: molecular architecture of mammalian polynucleotide kinase, a dna2 repair enzyme
|
| 78 | c2yy8B_ |
|
not modelled |
7.0 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:upf0106 protein ph0461;
PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
|
| 79 | d1sota1 |
|
not modelled |
6.9 |
18 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 80 | d1h3za_ |
|
not modelled |
6.9 |
25 |
Fold:SH3-like barrel
Superfamily:Tudor/PWWP/MBT
Family:PWWP domain |
| 81 | d1cz5a1 |
|
not modelled |
6.8 |
38 |
Fold:Double psi beta-barrel
Superfamily:ADC-like
Family:Cdc48 N-terminal domain-like |
| 82 | d1knwa1 |
|
not modelled |
6.8 |
20 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 83 | c1i7oC_ |
|
not modelled |
6.8 |
17 |
PDB header:isomerase, lyase
Chain: C: PDB Molecule:4-hydroxyphenylacetate degradation bifunctional
PDBTitle: crystal structure of hpce
|
| 84 | d1d7ka1 |
|
not modelled |
6.7 |
21 |
Fold:Domain of alpha and beta subunits of F1 ATP synthase-like
Superfamily:Alanine racemase C-terminal domain-like
Family:Eukaryotic ODC-like |
| 85 | c2hc8A_ |
|
not modelled |
6.6 |
33 |
PDB header:transport protein
Chain: A: PDB Molecule:cation-transporting atpase, p-type;
PDBTitle: structure of the a. fulgidus copa a-domain
|
| 86 | c4a1bO_ |
|
not modelled |
6.5 |
24 |
PDB header:ribosome
Chain: O: PDB Molecule:rpl28;
PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 26s rrna and3 proteins of molecule 3.
|
| 87 | c2p3wB_ |
|
not modelled |
6.4 |
27 |
PDB header:protein binding
Chain: B: PDB Molecule:probable serine protease htra3;
PDBTitle: crystal structure of the htra3 pdz domain bound to a phage-derived2 ligand (fgrwv)
|
| 88 | c3c9qA_ |
|
not modelled |
6.4 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein c8orf32;
PDBTitle: crystal structure of the uncharacterized human protein c8orf32 with2 bound peptide
|
| 89 | d1l0wa2 |
|
not modelled |
6.2 |
30 |
Fold:DCoH-like
Superfamily:GAD domain-like
Family:GAD domain |
| 90 | c3d3rA_ |
|
not modelled |
6.1 |
23 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
| 91 | d1clia2 |
|
not modelled |
6.1 |
13 |
Fold:PurM C-terminal domain-like
Superfamily:PurM C-terminal domain-like
Family:PurM C-terminal domain-like |
| 92 | d2eyqa1 |
|
not modelled |
6.1 |
35 |
Fold:SH3-like barrel
Superfamily:CarD-like
Family:CarD-like |
| 93 | d1vi6a_ |
|
not modelled |
6.1 |
14 |
Fold:Flavodoxin-like
Superfamily:Ribosomal protein S2
Family:Ribosomal protein S2 |
| 94 | c2qniA_ |
|
not modelled |
6.0 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein atu0299;
PDBTitle: crystal structure of uncharacterized protein atu0299
|
| 95 | c1yfsB_ |
|
not modelled |
5.9 |
22 |
PDB header:ligase
Chain: B: PDB Molecule:alanyl-trna synthetase;
PDBTitle: the crystal structure of alanyl-trna synthetase in complex2 with l-alanine
|
| 96 | d1ky9a1 |
|
not modelled |
5.9 |
25 |
Fold:PDZ domain-like
Superfamily:PDZ domain-like
Family:HtrA-like serine proteases |
| 97 | c2j5uB_ |
|
not modelled |
5.8 |
20 |
PDB header:cell shape regulation
Chain: B: PDB Molecule:mrec protein;
PDBTitle: mrec lysteria monocytogenes
|
| 98 | c2qq4A_ |
|
not modelled |
5.8 |
29 |
PDB header:metal binding protein
Chain: A: PDB Molecule:iron-sulfur cluster biosynthesis protein iscu;
PDBTitle: crystal structure of iron-sulfur cluster biosynthesis2 protein iscu (ttha1736) from thermus thermophilus hb8
|
| 99 | c2hw2A_ |
|
not modelled |
5.8 |
44 |
PDB header:transferase
Chain: A: PDB Molecule:rifampin adp-ribosyl transferase;
PDBTitle: crystal structure of rifampin adp-ribosyl transferase in2 complex with rifampin
|