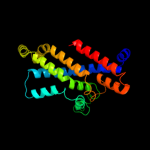
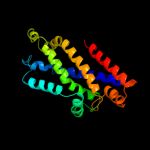
1 c2onkC_
99.9
15
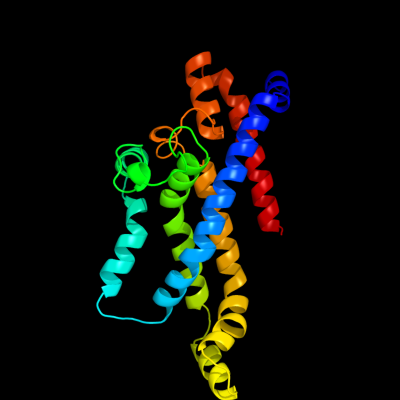
PDB header: membrane proteinChain: C: PDB Molecule: molybdate/tungstate abc transporter, permeasePDBTitle: abc transporter modbc in complex with its binding protein2 moda
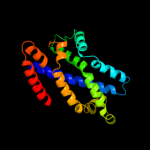
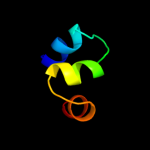
2 d2onkc1
99.9
15
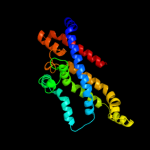
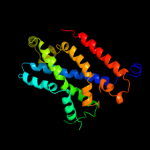
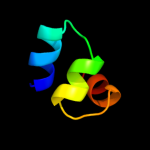
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like3 d3d31c1
99.9
14
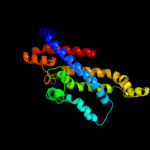
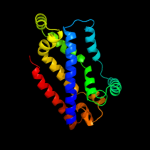
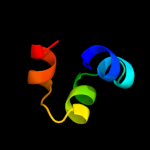
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like4 c3d31D_
99.9
14
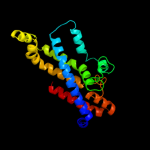
PDB header: transport proteinChain: D: PDB Molecule: sulfate/molybdate abc transporter, permeasePDBTitle: modbc from methanosarcina acetivorans
5 c2r6gF_
99.8
13
PDB header: hydrolase/transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: the crystal structure of the e. coli maltose transporter
6 d2r6gf2
99.8
14
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like7 c3fh6F_
99.8
12
PDB header: transport proteinChain: F: PDB Molecule: maltose transport system permease protein malf;PDBTitle: crystal structure of the resting state maltose transporter from e.2 coli
8 d2r6gg1
99.7
14
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like9 d3dhwa1
99.7
14
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like10 d1ntca_
35.3
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like11 d1umqa_
34.0
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like12 c1umqA_
34.0
22
PDB header: dna-binding proteinChain: A: PDB Molecule: photosynthetic apparatus regulatory protein;PDBTitle: solution structure and dna binding of the effector domain2 from the global regulator prra(rega) from r. sphaeroides:3 insights into dna binding specificity
13 d1etob_
32.3
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like14 d1fipa_
32.0
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like15 d1etxa_
27.0
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like16 c3e7lD_
24.1
13
PDB header: transcription regulatorChain: D: PDB Molecule: transcriptional regulator (ntrc family);PDBTitle: crystal structure of sigma54 activator ntrc4's dna binding2 domain
17 c2jwaA_
23.1
6
PDB header: transferaseChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: erbb2 transmembrane segment dimer spatial structure
18 d1g2ha_
21.4
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: FIS-like19 d2ns0a1
18.3
17
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like20 c2cw1A_
17.8
43
PDB header: de novo proteinChain: A: PDB Molecule: sn4m;PDBTitle: solution structure of the de novo-designed lambda cro fold2 protein
21 c2hx6A_
not modelled
13.1
14
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease;PDBTitle: solution structure analysis of the phage t42 endoribonuclease regb
22 c2d7dB_
not modelled
11.3
15
PDB header: hydrolase/dnaChain: B: PDB Molecule: 40-mer from uvrabc system protein b;PDBTitle: structural insights into the cryptic dna dependent atp-ase2 activity of uvrb
23 d1i3ja_
not modelled
8.9
10
Fold: DNA-binding domain of intron-encoded endonucleasesSuperfamily: DNA-binding domain of intron-encoded endonucleasesFamily: DNA-binding domain of intron-encoded endonucleases24 c3qkbB_
not modelled
8.7
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function which belongs to2 pfam duf74 family (pepe_0654) from pediococcus pentosaceus atcc 257453 at 2.73 a resolution
25 c3iwcD_
not modelled
8.3
35
PDB header: lyaseChain: D: PDB Molecule: s-adenosylmethionine decarboxylase;PDBTitle: t. maritima adometdc complex with s-adenosylmethionine2 methyl ester
26 d1st6a6
not modelled
8.2
15
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin27 d2auwa1
not modelled
8.0
26
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: NE0471 C-terminal domain-like28 d1i1ga1
not modelled
7.9
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain29 d1x8da1
not modelled
7.9
7
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: YiiL-like30 c2kvlA_
not modelled
7.8
8
PDB header: viral proteinChain: A: PDB Molecule: major outer capsid protein vp7;PDBTitle: nmr structure of the c-terminal domain of vp7
31 d1cf7a_
not modelled
7.6
19
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Cell cycle transcription factor e2f-dp32 c1twcF_
not modelled
7.5
29
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iii 23PDBTitle: rna polymerase ii complexed with gtp
33 d1twff_
not modelled
7.5
29
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB634 c2lhuA_
not modelled
7.4
19
PDB header: structural proteinChain: A: PDB Molecule: mybpc3 protein;PDBTitle: structural insight into the unique cardiac myosin binding protein-c2 motif: a partially folded domain
35 d1qkla_
not modelled
7.3
29
Fold: RPB6/omega subunit-likeSuperfamily: RPB6/omega subunit-likeFamily: RPB636 c2pmzW_
not modelled
7.3
14
PDB header: translation, transferaseChain: W: PDB Molecule: dna-directed rna polymerase subunit k;PDBTitle: archaeal rna polymerase from sulfolobus solfataricus
37 c3s1bA_
not modelled
7.3
24
PDB header: signaling proteinChain: A: PDB Molecule: mini-z;PDBTitle: the development of peptide-based tools for the analysis of2 angiogenesis
38 c1ciiA_
not modelled
7.2
30
PDB header: transmembrane proteinChain: A: PDB Molecule: colicin ia;PDBTitle: colicin ia
39 d2cg4a1
not modelled
7.1
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain40 d3orca_
not modelled
7.0
36
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors41 d2cyya1
not modelled
7.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain42 c1y2iC_
not modelled
7.0
27
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein s0862;PDBTitle: crystal structure of mcsg target apc27401 from shigella2 flexneri
43 d1y2ia_
not modelled
7.0
27
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like44 c2ks1B_
not modelled
6.9
7
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
45 d2hqva1
not modelled
6.9
8
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: ChuX-like46 d2obpa1
not modelled
6.8
23
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: ReutB4095-like47 c2nq2A_
not modelled
6.8
11
PDB header: metal transportChain: A: PDB Molecule: hypothetical abc transporter permease proteinPDBTitle: an inward-facing conformation of a putative metal-chelate2 type abc transporter.
48 d1se7a_
not modelled
6.7
40
Fold: DNA polymerase III theta subunit-likeSuperfamily: DNA polymerase III theta subunit-likeFamily: DNA polymerase III theta subunit-like49 d2cfxa1
not modelled
6.6
6
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: Lrp/AsnC-like transcriptional regulator N-terminal domain50 c2qlxA_
not modelled
6.6
11
PDB header: isomeraseChain: A: PDB Molecule: l-rhamnose mutarotase;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
51 c2qlwA_
not modelled
6.6
11
PDB header: isomeraseChain: A: PDB Molecule: rhau;PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
52 d1vr4a1
not modelled
6.4
9
Fold: Dodecin subunit-likeSuperfamily: YbjQ-likeFamily: YbjQ-like53 d1sdia_
not modelled
6.4
23
Fold: YcfC-likeSuperfamily: YcfC-likeFamily: YcfC-like54 c2auwB_
not modelled
6.4
19
PDB header: unknown functionChain: B: PDB Molecule: hypothetical protein ne0471;PDBTitle: crystal structure of putative dna binding protein ne0471 from2 nitrosomonas europaea atcc 19718
55 c2zihC_
not modelled
6.4
6
PDB header: protein transportChain: C: PDB Molecule: vacuolar protein sorting-associated protein 74;PDBTitle: crystal structure of yeast vps74
56 d1v54g_
not modelled
6.4
12
Fold: Single transmembrane helixSuperfamily: Mitochondrial cytochrome c oxidase subunit VIaFamily: Mitochondrial cytochrome c oxidase subunit VIa57 d1brwa3
not modelled
6.4
24
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain58 c2i88A_
not modelled
6.4
24
PDB header: membrane proteinChain: A: PDB Molecule: colicin-e1;PDBTitle: crystal structure of the channel-forming domain of colicin2 e1
59 c2ziiA_
not modelled
6.3
6
PDB header: protein transportChain: A: PDB Molecule: vacuolar protein sorting-associated protein 74;PDBTitle: crystal structure of yeast vps74-n-term truncation variant
60 d2o3aa1
not modelled
6.3
33
Fold: alpha/beta knotSuperfamily: alpha/beta knotFamily: AF0751-like61 d1o8ca2
not modelled
6.3
15
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain62 c2kz3A_
not modelled
6.3
18
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein rad51l3;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for human rad51d2 from 1 to 83
63 d1h6gb1
not modelled
6.3
19
Fold: Four-helical up-and-down bundleSuperfamily: alpha-catenin/vinculin-likeFamily: alpha-catenin/vinculin64 d2phcb1
not modelled
6.2
8
Fold: Cyclophilin-likeSuperfamily: Cyclophilin-likeFamily: PH0987 C-terminal domain-like65 c2yy8B_
not modelled
6.2
33
PDB header: transferaseChain: B: PDB Molecule: upf0106 protein ph0461;PDBTitle: crystal structure of archaeal trna-methylase for position2 56 (atrm56) from pyrococcus horikoshii, complexed with s-3 adenosyl-l-methionine
66 d2tpta3
not modelled
6.2
29
Fold: alpha/beta-HammerheadSuperfamily: Pyrimidine nucleoside phosphorylase C-terminal domainFamily: Pyrimidine nucleoside phosphorylase C-terminal domain67 c1pyuD_
not modelled
6.1
31
PDB header: lyaseChain: D: PDB Molecule: aspartate 1-decarboxylase alfa chain;PDBTitle: processed aspartate decarboxylase mutant with ser25 mutated to cys
68 c2vn2B_
not modelled
6.1
14
PDB header: replicationChain: B: PDB Molecule: chromosome replication initiation protein;PDBTitle: crystal structure of the n-terminal domain of dnad protein2 from geobacillus kaustophilus hta426
69 c1piiA_
not modelled
6.0
20
PDB header: bifunctional(isomerase and synthase)Chain: A: PDB Molecule: n-(5'phosphoribosyl)anthranilate isomerase;PDBTitle: three-dimensional structure of the bifunctional enzyme2 phosphoribosylanthranilate isomerase:3 indoleglycerolphosphate synthase from escherichia coli4 refined at 2.0 angstroms resolution
70 c1rkcB_
not modelled
6.0
11
PDB header: cell adhesion, structural proteinChain: B: PDB Molecule: talin;PDBTitle: human vinculin head (1-258) in complex with talin's2 vinculin binding site 3 (residues 1944-1969)
71 d1jhfa1
not modelled
6.0
12
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: LexA repressor, N-terminal DNA-binding domain72 c2gqqB_
not modelled
6.0
6
PDB header: transcriptionChain: B: PDB Molecule: leucine-responsive regulatory protein;PDBTitle: crystal structure of e. coli leucine-responsive regulatory protein2 (lrp)
73 c3mmlD_
not modelled
5.9
17
PDB header: hydrolaseChain: D: PDB Molecule: allophanate hydrolase subunit 1;PDBTitle: allophanate hydrolase complex from mycobacterium smegmatis, msmeg0435-2 msmeg0436
74 c3h0gF_
not modelled
5.9
27
PDB header: transcriptionChain: F: PDB Molecule: dna-directed rna polymerases i, ii, and iiiPDBTitle: rna polymerase ii from schizosaccharomyces pombe
75 c1xwjB_
not modelled
5.9
11
PDB header: cell adhesion/protein bindingChain: B: PDB Molecule: talin;PDBTitle: vinculin head (1-258) in complex with the talin vinculin2 binding site 3 (1945-1969)
76 d1icha_
not modelled
5.9
5
Fold: DEATH domainSuperfamily: DEATH domainFamily: DEATH domain, DD77 c1ichA_
not modelled
5.9
5
PDB header: apoptosisChain: A: PDB Molecule: tumor necrosis factor receptor-1;PDBTitle: solution structure of the tumor necrosis factor receptor-12 death domain
78 c2ka1A_
not modelled
5.8
11
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
79 c2ka2B_
not modelled
5.8
11
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
80 d1a53a_
not modelled
5.8
27
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes81 c2e75E_
not modelled
5.8
29
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with 2-nonyl-4-2 hydroxyquinoline n-oxide (nqno) from m.laminosus
82 c1vf5R_
not modelled
5.8
29
PDB header: photosynthesisChain: R: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus
83 c2e74E_
not modelled
5.8
29
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex from m.laminosus
84 c2e76E_
not modelled
5.8
29
PDB header: photosynthesisChain: E: PDB Molecule: cytochrome b6-f complex subunit 6;PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
85 d2e74e1
not modelled
5.8
29
Fold: Single transmembrane helixSuperfamily: PetL subunit of the cytochrome b6f complexFamily: PetL subunit of the cytochrome b6f complex86 c2xa6B_
not modelled
5.7
21
PDB header: transcriptionChain: B: PDB Molecule: kh domain-containing\,rna-binding\,signalPDBTitle: structural basis for homodimerization of the src-associated2 during mitosis, 68 kd protein (sam68) qua1 domain
87 c2ka2A_
not modelled
5.7
16
PDB header: membrane proteinChain: A: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles with his173-ser172 intermonomer3 hydrogen bond restraints
88 c2ka1B_
not modelled
5.7
16
PDB header: membrane proteinChain: B: PDB Molecule: bcl2/adenovirus e1b 19 kda protein-interactingPDBTitle: solution nmr structure of bnip3 transmembrane peptide dimer2 in detergent micelles
89 c3lpeF_
not modelled
5.7
25
PDB header: transferaseChain: F: PDB Molecule: dna-directed rna polymerase subunit e'';PDBTitle: crystal structure of spt4/5ngn heterodimer complex from methanococcus2 jannaschii
90 d1nyoa_
not modelled
5.7
3
Fold: FAS1 domainSuperfamily: FAS1 domainFamily: FAS1 domain91 c2ijlB_
not modelled
5.6
20
PDB header: transcriptionChain: B: PDB Molecule: molybdenum-binding transcriptional repressor;PDBTitle: the structure of a putative mode from agrobacterium tumefaciens.
92 d1vf5c3
not modelled
5.6
17
Fold: Single transmembrane helixSuperfamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchorFamily: Cytochrome f subunit of the cytochrome b6f complex, transmembrane anchor93 c2zp2B_
not modelled
5.5
4
PDB header: transferase inhibitorChain: B: PDB Molecule: kinase a inhibitor;PDBTitle: c-terminal domain of kipi from bacillus subtilis
94 d1l7va_
not modelled
5.5
13
Fold: ABC transporter involved in vitamin B12 uptake, BtuCSuperfamily: ABC transporter involved in vitamin B12 uptake, BtuCFamily: ABC transporter involved in vitamin B12 uptake, BtuC95 c2kncA_
not modelled
5.5
16
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
96 c1jd7A_
not modelled
5.5
13
PDB header: hydrolaseChain: A: PDB Molecule: alpha-amylase;PDBTitle: crystal structure analysis of the mutant k300r of2 pseudoalteromonas haloplanctis alpha-amylase
97 c3gebC_
not modelled
5.4
17
PDB header: hydrolaseChain: C: PDB Molecule: eyes absent homolog 2;PDBTitle: crystal structure of edeya2
98 c3gk0H_
not modelled
5.4
27
PDB header: transferaseChain: H: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: crystal structure of pyridoxal phosphate biosynthetic2 protein from burkholderia pseudomallei
99 c1vf5E_
not modelled
5.4
30
PDB header: photosynthesisChain: E: PDB Molecule: protein pet l;PDBTitle: crystal structure of cytochrome b6f complex from m.laminosus