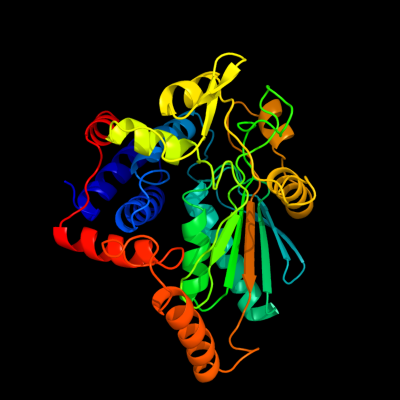
| 1 | d1nxua_
|
|
 |
100.0 |
95 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
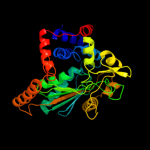
| 2 | c1vbiA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:type 2 malate/lactate dehydrogenase;
PDBTitle: crystal structure of type 2 malate/lactate dehydrogenase from thermus2 thermophilus hb8
|
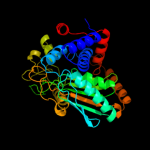
| 3 | d1xrha_
|
|
 |
100.0 |
22 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
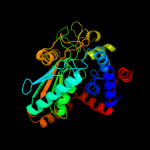
| 4 | c3i0pA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of malate dehydrogenase from entamoeba histolytica
|
| 5 | c1z2iA_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of agrobacterium tumefaciens malate2 dehydrogenase, new york structural genomics consortium
|
| 6 | d1rfma_
|
|
 |
100.0 |
27 |
Fold:L-sulfolactate dehydrogenase-like
Superfamily:L-sulfolactate dehydrogenase-like
Family:L-sulfolactate dehydrogenase-like |
| 7 | c1wtjB_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ureidoglycolate dehydrogenase;
PDBTitle: crystal structure of delta1-piperideine-2-carboxylate2 reductase from pseudomonas syringae pvar.tomato
|
| 8 | c2g8yB_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate/l-lactate dehydrogenases;
PDBTitle: the structure of a putative malate/lactate dehydrogenase from e. coli.
|
| 9 | c1v9nA_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: structure of malate dehydrogenase from pyrococcus horikoshii ot3
|
| 10 | c3uoeB_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dehydrogenase;
PDBTitle: the crystal structure of dehydrogenase from sinorhizobium meliloti
|
| 11 | c3eefA_
|
|
 |
49.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-carbamoylsarcosine amidase related protein;
PDBTitle: crystal structure of n-carbamoylsarcosine amidase from thermoplasma2 acidophilum
|
| 12 | c2qiwA_
|
|
 |
48.9 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
| 13 | c3irvA_
|
|
 |
46.7 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:cysteine hydrolase;
PDBTitle: crystal structure of cysteine hydrolase pspph_2384 from pseudomonas2 syringae pv. phaseolicola 1448a
|
| 14 | c3kl2K_
|
|
 |
46.2 |
11 |
PDB header:structural genomics, unknown function
Chain: K: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase from streptomyces2 avermitilis
|
| 15 | c2ze3A_
|
|
 |
42.9 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
| 16 | d1muma_
|
|
 |
41.7 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 17 | c2hjpA_
|
|
 |
38.3 |
8 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
| 18 | d1nbaa_
|
|
 |
37.7 |
20 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 19 | d2ebfx2
|
|
 |
37.3 |
25 |
Fold:EreA/ChaN-like
Superfamily:EreA/ChaN-like
Family:PMT domain-like |
| 20 | c3b8iF_
|
|
 |
37.3 |
14 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
| 21 | c2b34C_ |
|
not modelled |
37.3 |
17 |
PDB header:hydrolase
Chain: C: PDB Molecule:mar1 ribonuclease;
PDBTitle: structure of mar1 ribonuclease from caenorhabditis elegans
|
| 22 | d1nf9a_ |
|
not modelled |
37.2 |
11 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 23 | c3ih1A_ |
|
not modelled |
36.2 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
| 24 | c3ot4F_ |
|
not modelled |
35.7 |
11 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative isochorismatase;
PDBTitle: structure and catalytic mechanism of bordetella bronchiseptica nicf
|
| 25 | c3eooL_ |
|
not modelled |
31.8 |
13 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
| 26 | c1e0mA_ |
|
not modelled |
28.8 |
21 |
PDB header:de novo protein
Chain: A: PDB Molecule:wwprototype;
PDBTitle: prototype ww domain
|
| 27 | c1yiuA_ |
|
not modelled |
27.4 |
29 |
PDB header:ligase
Chain: A: PDB Molecule:itchy e3 ubiquitin protein ligase;
PDBTitle: itch e3 ubiquitin ligase ww3 domain
|
| 28 | c2ysbA_ |
|
not modelled |
25.9 |
18 |
PDB header:protein binding
Chain: A: PDB Molecule:salvador homolog 1 protein;
PDBTitle: solution structure of the first ww domain from the mouse2 salvador homolog 1 protein (sav1)
|
| 29 | d1wdjb_ |
|
not modelled |
25.4 |
10 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Hypothetical protein TT1808 (TTHA1514) |
| 30 | c3mcwA_ |
|
not modelled |
25.3 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative hydrolase;
PDBTitle: crystal structure of an a putative hydrolase of the isochorismatase2 family (cv_1320) from chromobacterium violaceum atcc 12472 at 1.06 a3 resolution
|
| 31 | c3l4hA_ |
|
not modelled |
25.2 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase hecw1;
PDBTitle: helical box domain and second ww domain of the human e3 ubiquitin-2 protein ligase hecw1
|
| 32 | c3hb7G_ |
|
not modelled |
24.6 |
16 |
PDB header:hydrolase
Chain: G: PDB Molecule:isochorismatase hydrolase;
PDBTitle: the crystal structure of an isochorismatase-like hydrolase from2 alkaliphilus metalliredigens to 2.3a
|
| 33 | d1wdja_ |
|
not modelled |
23.9 |
10 |
Fold:Restriction endonuclease-like
Superfamily:Restriction endonuclease-like
Family:Hypothetical protein TT1808 (TTHA1514) |
| 34 | c2px0D_ |
|
not modelled |
23.0 |
10 |
PDB header:biosynthetic protein
Chain: D: PDB Molecule:flagellar biosynthesis protein flhf;
PDBTitle: crystal structure of flhf complexed with gmppnp/mg(2+)
|
| 35 | c2ysdA_ |
|
not modelled |
22.9 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:membrane-associated guanylate kinase, ww and pdz
PDBTitle: solution structure of the first ww domain from the human2 membrane-associated guanylate kinase, ww and pdz domain-3 containing protein 1. magi-1
|
| 36 | d1f8ab1 |
|
not modelled |
22.8 |
20 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 37 | d1yaca_ |
|
not modelled |
22.4 |
15 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 38 | c1wr7A_ |
|
not modelled |
22.2 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:nedd4-2;
PDBTitle: solution structure of the third ww domain of nedd4-2
|
| 39 | c2fq1A_ |
|
not modelled |
21.6 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:isochorismatase;
PDBTitle: crystal structure of the two-domain non-ribosomal peptide synthetase2 entb containing isochorismate lyase and aryl-carrier protein domains
|
| 40 | d1sgma2 |
|
not modelled |
21.4 |
25 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
| 41 | c2djyA_ |
|
not modelled |
21.4 |
21 |
PDB header:ligase/signaling protein
Chain: A: PDB Molecule:smad ubiquitination regulatory factor 2;
PDBTitle: solution structure of smurf2 ww3 domain-smad7 py peptide2 complex
|
| 42 | c3ot2A_ |
|
not modelled |
21.2 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative nuclease belonging to duf8202 (ava_3926) from anabaena variabilis atcc 29413 at 1.96 a resolution
|
| 43 | c3ot2B_ |
|
not modelled |
21.2 |
10 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative nuclease belonging to duf8202 (ava_3926) from anabaena variabilis atcc 29413 at 1.96 a resolution
|
| 44 | d1t57a_ |
|
not modelled |
20.9 |
16 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 45 | c1wr4A_ |
|
not modelled |
20.7 |
29 |
PDB header:ligase
Chain: A: PDB Molecule:ubiquitin-protein ligase nedd4-2;
PDBTitle: solution structure of the second ww domain of nedd4-2
|
| 46 | d1bdga1 |
|
not modelled |
20.5 |
16 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
| 47 | c2lawA_ |
|
not modelled |
20.4 |
21 |
PDB header:signaling protein/transcription
Chain: A: PDB Molecule:yorkie homolog;
PDBTitle: structure of the second ww domain from human yap in complex with a2 human smad1 derived peptide
|
| 48 | c3oqpA_ |
|
not modelled |
20.3 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 49 | c1wmvA_ |
|
not modelled |
20.3 |
29 |
PDB header:oxidoreductase, apoptosis
Chain: A: PDB Molecule:ww domain containing oxidoreductase;
PDBTitle: solution structure of the second ww domain of wwox
|
| 50 | c2uygF_ |
|
not modelled |
20.0 |
20 |
PDB header:lyase
Chain: F: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystallogaphic structure of the typeii 3-dehydroquinase2 from thermus thermophilus
|
| 51 | d1k9ra_ |
|
not modelled |
19.8 |
18 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 52 | c2h0rD_ |
|
not modelled |
19.5 |
14 |
PDB header:hydrolase
Chain: D: PDB Molecule:nicotinamidase;
PDBTitle: structure of the yeast nicotinamidase pnc1p
|
| 53 | d2cfua2 |
|
not modelled |
19.5 |
14 |
Fold:Metallo-hydrolase/oxidoreductase
Superfamily:Metallo-hydrolase/oxidoreductase
Family:Alkylsulfatase-like |
| 54 | d3bzra1 |
|
not modelled |
19.4 |
13 |
Fold:EscU C-terminal domain-like
Superfamily:EscU C-terminal domain-like
Family:EscU C-terminal domain-like |
| 55 | c3bzrA_ |
|
not modelled |
19.4 |
13 |
PDB header:membrane protein, protein transport
Chain: A: PDB Molecule:escu;
PDBTitle: crystal structure of escu c-terminal domain with n262d mutation, space2 group p 41 21 2
|
| 56 | c3t7yB_ |
|
not modelled |
19.0 |
16 |
PDB header:protein transport
Chain: B: PDB Molecule:yop proteins translocation protein u;
PDBTitle: structure of an autocleavage-inactive mutant of the cytoplasmic domain2 of ct091, the yscu homologue of chlamydia trachomatis
|
| 57 | d3ci0k2 |
|
not modelled |
18.6 |
15 |
Fold:SAM domain-like
Superfamily:GspK insert domain-like
Family:GspK insert domain-like |
| 58 | d1tk7a1 |
|
not modelled |
18.3 |
14 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 59 | c1ymzA_ |
|
not modelled |
18.1 |
36 |
PDB header:unknown function
Chain: A: PDB Molecule:cc45;
PDBTitle: cc45, an artificial ww domain designed using statistical2 coupling analysis
|
| 60 | d1j2ra_ |
|
not modelled |
17.0 |
14 |
Fold:Isochorismatase-like hydrolases
Superfamily:Isochorismatase-like hydrolases
Family:Isochorismatase-like hydrolases |
| 61 | c2k29A_ |
|
not modelled |
16.5 |
33 |
PDB header:transcription
Chain: A: PDB Molecule:antitoxin relb;
PDBTitle: structure of the dbd domain of e. coli antitoxin relb
|
| 62 | d1i5hw_ |
|
not modelled |
16.5 |
20 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 63 | d1f61a_ |
|
not modelled |
16.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
| 64 | d3djba1 |
|
not modelled |
15.6 |
14 |
Fold:HD-domain/PDEase-like
Superfamily:HD-domain/PDEase-like
Family:HD domain |
| 65 | c3lqyA_ |
|
not modelled |
15.3 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative isochorismatase hydrolase;
PDBTitle: crystal structure of putative isochorismatase hydrolase from2 oleispira antarctica
|
| 66 | c2kxqA_ |
|
not modelled |
14.2 |
20 |
PDB header:protein binding
Chain: A: PDB Molecule:e3 ubiquitin-protein ligase smurf2;
PDBTitle: solution structure of smurf2 ww2 and ww3 bound to smad7 py motif2 containing peptide
|
| 67 | d1mnta_ |
|
not modelled |
14.0 |
19 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:Arc/Mnt-like phage repressors |
| 68 | d1a4sa_ |
|
not modelled |
13.9 |
18 |
Fold:ALDH-like
Superfamily:ALDH-like
Family:ALDH-like |
| 69 | c2jlhA_ |
|
not modelled |
13.7 |
16 |
PDB header:protein transport
Chain: A: PDB Molecule:yop proteins translocation protein u;
PDBTitle: crystal structure of the cytoplasmic domain of yersinia2 pestis yscu n263a mutant
|
| 70 | c3o93A_ |
|
not modelled |
13.0 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:nicotinamidase;
PDBTitle: high resolution crystal structures of streptococcus pneumoniae2 nicotinamidase with trapped intermediates provide insights into3 catalytic mechanism and inhibition by aldehydes
|
| 71 | c3hu5B_ |
|
not modelled |
13.0 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein from desulfovibrio2 vulgaris subsp. vulgaris str. hildenborough
|
| 72 | c3oqpB_ |
|
not modelled |
12.9 |
24 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative isochorismatase;
PDBTitle: crystal structure of a putative isochorismatase (bxe_a0706) from2 burkholderia xenovorans lb400 at 1.22 a resolution
|
| 73 | c3ivlA_ |
|
not modelled |
12.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative zinc protease;
PDBTitle: the crystal structure of the inactive peptidase domain of a putative2 zinc protease from bordetella parapertussis to 2.2a
|
| 74 | c2hg2A_ |
|
not modelled |
12.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde dehydrogenase a;
PDBTitle: structure of lactaldehyde dehydrogenase
|
| 75 | c3fpvC_ |
|
not modelled |
12.6 |
21 |
PDB header:heme binding protein
Chain: C: PDB Molecule:extracellular haem-binding protein;
PDBTitle: crystal structure of hbps
|
| 76 | d1lvaa3 |
|
not modelled |
12.5 |
11 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:C-terminal fragment of elongation factor SelB |
| 77 | d1zk8a2 |
|
not modelled |
12.5 |
21 |
Fold:Tetracyclin repressor-like, C-terminal domain
Superfamily:Tetracyclin repressor-like, C-terminal domain
Family:Tetracyclin repressor-like, C-terminal domain |
| 78 | c3le4A_ |
|
not modelled |
12.1 |
22 |
PDB header:nuclear protein
Chain: A: PDB Molecule:microprocessor complex subunit dgcr8;
PDBTitle: crystal structure of the dgcr8 dimerization domain
|
| 79 | c2dmvA_ |
|
not modelled |
11.9 |
27 |
PDB header:ligase
Chain: A: PDB Molecule:itchy homolog e3 ubiquitin protein ligase;
PDBTitle: solution structure of the second ww domain of itchy homolog2 e3 ubiquitin protein ligase (itch)
|
| 80 | c2h6rG_ |
|
not modelled |
11.8 |
18 |
PDB header:isomerase
Chain: G: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase (tim) from2 methanocaldococcus jannaschii
|
| 81 | c2dclB_ |
|
not modelled |
11.8 |
14 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:hypothetical upf0166 protein ph1503;
PDBTitle: structure of ph1503 protein from pyrococcus horikoshii ot3
|
| 82 | c3ju8B_ |
|
not modelled |
11.6 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:succinylglutamic semialdehyde dehydrogenase;
PDBTitle: crystal structure of succinylglutamic semialdehyde dehydrogenase from2 pseudomonas aeruginosa.
|
| 83 | c2a67C_ |
|
not modelled |
11.4 |
14 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:isochorismatase family protein;
PDBTitle: crystal structure of isochorismatase family protein
|
| 84 | c2cfuA_ |
|
not modelled |
11.4 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:sdsa1;
PDBTitle: crystal structure of sdsa1, an alkylsulfatase from2 pseudomonas aeruginosa, in complex with 1-decane-sulfonic-3 acid.
|
| 85 | c3k2wD_ |
|
not modelled |
11.3 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:betaine-aldehyde dehydrogenase;
PDBTitle: crystal structure of betaine-aldehyde dehydrogenase from2 pseudoalteromonas atlantica t6c
|
| 86 | d2jmfa1 |
|
not modelled |
11.3 |
21 |
Fold:WW domain-like
Superfamily:WW domain
Family:WW domain |
| 87 | d1o51a_ |
|
not modelled |
10.6 |
14 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:DUF190/COG1993 |
| 88 | c2xetB_ |
|
not modelled |
10.5 |
5 |
PDB header:transport protein
Chain: B: PDB Molecule:f1 capsule-anchoring protein;
PDBTitle: conserved hydrophobic clusters on the surface of the caf1a usher2 c-terminal domain are important for f1 antigen assembly
|
| 89 | c3e0kA_ |
|
not modelled |
10.5 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:amino-acid acetyltransferase;
PDBTitle: crystal structure of c-termianl domain of n-acetylglutamate synthase2 from vibrio parahaemolyticus
|
| 90 | d2bfda1 |
|
not modelled |
10.5 |
11 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:Branched-chain alpha-keto acid dehydrogenase PP module |
| 91 | c2vovA_ |
|
not modelled |
10.4 |
38 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:surface-associated protein;
PDBTitle: an oxidized tryptophan facilitates copper-binding in2 methylococcus capsulatus secreted protein mope. the3 stucture of wild-type mope to 1.35aa
|
| 92 | c3jz4C_ |
|
not modelled |
10.3 |
26 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:succinate-semialdehyde dehydrogenase [nadp+];
PDBTitle: crystal structure of e. coli nadp dependent enzyme
|
| 93 | c3iwkB_ |
|
not modelled |
10.3 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:aminoaldehyde dehydrogenase;
PDBTitle: crystal structure of aminoaldehyde dehydrogenase 1 from2 pisum sativum (psamadh1)
|
| 94 | d1vp8a_ |
|
not modelled |
10.3 |
19 |
Fold:Pyruvate kinase C-terminal domain-like
Superfamily:PK C-terminal domain-like
Family:MTH1675-like |
| 95 | c1qzrA_ |
|
not modelled |
10.3 |
10 |
PDB header:isomerase
Chain: A: PDB Molecule:dna topoisomerase ii;
PDBTitle: crystal structure of the atpase region of saccharomyces cerevisiae2 topoisomerase ii bound to icrf-187 (dexrazoxane)
|
| 96 | c3hazA_ |
|
not modelled |
10.1 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:proline dehydrogenase;
PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
|
| 97 | d1nnsa_ |
|
not modelled |
10.0 |
14 |
Fold:Glutaminase/Asparaginase
Superfamily:Glutaminase/Asparaginase
Family:Glutaminase/Asparaginase |
| 98 | d2pw9a1 |
|
not modelled |
10.0 |
26 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:FdhD/NarQ |
| 99 | d1ig8a1 |
|
not modelled |
10.0 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |