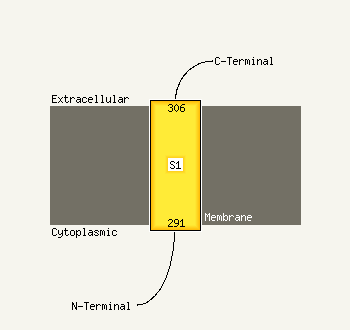
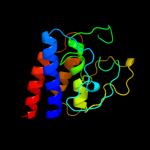
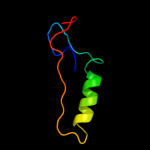
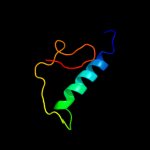
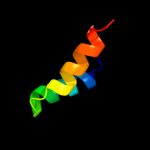
1 d2ppqa1
90.7
13
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases2 d1zyla1
84.1
10
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases3 c3dxqB_
79.8
18
PDB header: transferaseChain: B: PDB Molecule: choline/ethanolamine kinase family protein;PDBTitle: crystal structure of choline/ethanolamine kinase family protein2 (np_106042.1) from mesorhizobium loti at 2.55 a resolution
4 d1nd4a_
67.5
15
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases5 c3akjB_
58.5
9
PDB header: transferaseChain: B: PDB Molecule: ctka;PDBTitle: crystal structure of a helicobacter pylori proinflammatory kinase ctka
6 c3ovcA_
50.2
13
PDB header: transferase/antibioticChain: A: PDB Molecule: hygromycin-b 4-o-kinase;PDBTitle: crystal structure of aminoglycoside phosphotransferase aph(4)-ia
7 c2j0kB_
47.1
13
PDB header: transferaseChain: B: PDB Molecule: focal adhesion kinase 1;PDBTitle: crystal structure of a fragment of focal adhesion kinase2 containing the ferm and kinase domains.
8 c1u0bB_
40.6
24
PDB header: ligase/rnaChain: B: PDB Molecule: cysteinyl trna;PDBTitle: crystal structure of cysteinyl-trna synthetase binary2 complex with trnacys
9 c1zp9A_
40.1
24
PDB header: transferaseChain: A: PDB Molecule: rio1 kinase;PDBTitle: crystal structure of full-legnth a.fulgidus rio1 serine kinase bound2 to atp and mn2+ ions.
10 c2vzaD_
39.6
25
PDB header: cell adhesionChain: D: PDB Molecule: cell filamentation protein;PDBTitle: type iv secretion system effector protein bepa
11 d1dx4a_
39.5
23
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like12 c1unhD_
37.9
29
PDB header: cell cycleChain: D: PDB Molecule: cyclin-dependent kinase 5 activator 1;PDBTitle: structural mechanism for the inhibition of cdk5-p25 by2 roscovitine, aloisine and indirubin.
13 d1unld_
37.6
29
Fold: Cyclin-likeSuperfamily: Cyclin-likeFamily: Cyclin14 d2i8da1
37.2
13
Fold: Secretion chaperone-likeSuperfamily: YdhG-likeFamily: YdhG-like15 d1zara2
34.9
18
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: RIO1-like kinases16 c3dxpA_
34.1
11
PDB header: transferaseChain: A: PDB Molecule: putative acyl-coa dehydrogenase;PDBTitle: crystal structure of a putative aminoglycoside phosphotransferase2 (reut_a1007) from ralstonia eutropha jmp134 at 2.32 a resolution
17 d1dgja4
34.0
23
Fold: Molybdenum cofactor-binding domainSuperfamily: Molybdenum cofactor-binding domainFamily: Molybdenum cofactor-binding domain18 d1p0ia_
33.0
22
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like19 d2f2ab1
32.3
18
Fold: GatB/YqeY motifSuperfamily: GatB/YqeY motifFamily: GatB/GatE C-terminal domain-like20 c3e7eA_
31.5
21
PDB header: transferaseChain: A: PDB Molecule: mitotic checkpoint serine/threonine-protein kinase bub1;PDBTitle: structure and substrate recruitment of the human spindle checkpoint2 kinase bub
21 c2pm8A_
not modelled
31.0
22
PDB header: hydrolaseChain: A: PDB Molecule: cholinesterase;PDBTitle: crystal structure of recombinant full length human2 butyrylcholinesterase
22 c1vlbA_
not modelled
31.0
18
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde oxidoreductase;PDBTitle: structure refinement of the aldehyde oxidoreductase from2 desulfovibrio gigas at 1.28 a
23 c3k1sE_
not modelled
27.9
23
PDB header: transferaseChain: E: PDB Molecule: pts system, cellobiose-specific iia component;PDBTitle: crystal structure of the pts cellobiose specific enzyme iia2 from bacillus anthracis
24 c1q5vB_
not modelled
26.5
28
PDB header: transcriptionChain: B: PDB Molecule: nickel responsive regulator;PDBTitle: apo-nikr
25 d1dfoa_
not modelled
25.4
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like26 c3l8rA_
not modelled
24.1
9
PDB header: transferaseChain: A: PDB Molecule: putative pts system, cellobiose-specific iiaPDBTitle: the crystal structure of ptca from s. mutans
27 c3eu8D_
not modelled
23.5
38
PDB header: hydrolaseChain: D: PDB Molecule: putative glucoamylase;PDBTitle: crystal structure of putative glucoamylase (yp_210071.1) from2 bacteroides fragilis nctc 9343 at 2.12 a resolution
28 d1j7la_
not modelled
23.2
27
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases29 c1wcrA_
not modelled
23.1
27
PDB header: transferaseChain: A: PDB Molecule: pts system, n, n'-diacetylchitobiose-specificPDBTitle: trimeric structure of the enzyme iia from escherichia coli2 phosphotransferase system specific for n,n'-3 diacetylchitobiose
30 d2e2aa_
not modelled
22.5
32
Fold: Spectrin repeat-likeSuperfamily: Enzyme IIa from lactose specific PTS, IIa-lacFamily: Enzyme IIa from lactose specific PTS, IIa-lac31 d2ha2a1
not modelled
22.4
26
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like32 d1vlya1
not modelled
22.2
21
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain33 c2zqeA_
not modelled
22.0
44
PDB header: dna binding proteinChain: A: PDB Molecule: muts2 protein;PDBTitle: crystal structure of the smr domain of thermus thermophilus muts2
34 d1bwva2
not modelled
21.9
17
Fold: Ferredoxin-likeSuperfamily: RuBisCO, large subunit, small (N-terminal) domainFamily: Ribulose 1,5-bisphosphate carboxylase-oxygenase35 c2xm5A_
not modelled
21.5
21
PDB header: transferaseChain: A: PDB Molecule: cloq;PDBTitle: structural and mechanistic analysis of the magnesium-2 independent aromatic prenyltransferase cloq from the3 clorobiocin biosynthetic pathway
36 d2burb1
not modelled
21.2
25
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase37 d1g8fa3
not modelled
21.1
26
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: ATP sulfurylase C-terminal domain38 d1qe3a_
not modelled
20.2
27
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like39 d1wdka2
not modelled
20.0
13
Fold: 6-phosphogluconate dehydrogenase C-terminal domain-likeSuperfamily: 6-phosphogluconate dehydrogenase C-terminal domain-likeFamily: HCDH C-domain-like40 d1f6wa_
not modelled
19.4
29
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like41 d1tzva_
not modelled
19.3
6
Fold: NusB-likeSuperfamily: NusB-likeFamily: Antitermination factor NusB42 c3f7wA_
not modelled
18.9
14
PDB header: transferaseChain: A: PDB Molecule: putative fructosamine-3-kinase;PDBTitle: crystal structure of putative fructosamine-3-kinase (yp_290396.1) from2 thermobifida fusca yx-er1 at 1.85 a resolution
43 d2oc6a1
not modelled
18.8
11
Fold: Secretion chaperone-likeSuperfamily: YdhG-likeFamily: YdhG-like44 c3attA_
not modelled
18.7
13
PDB header: transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of rv3168 with atp
45 c3h0mE_
not modelled
18.6
12
PDB header: ligaseChain: E: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferasePDBTitle: structure of trna-dependent amidotransferase gatcab from2 aquifex aeolicus
46 d2apla1
not modelled
18.2
11
Fold: PG0816-likeSuperfamily: PG0816-likeFamily: PG0816-like47 c3al0B_
not modelled
18.1
15
PDB header: ligase/rnaChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferase subunit b;PDBTitle: crystal structure of the glutamine transamidosome from thermotoga2 maritima in the glutamylation state.
48 c2ogsA_
not modelled
17.9
22
PDB header: hydrolaseChain: A: PDB Molecule: thermostable carboxylesterase est50;PDBTitle: crystal structure of the geobacillus stearothermophilus2 carboxylesterase est55 at ph 6.2
49 c3en9B_
not modelled
17.7
15
PDB header: hydrolaseChain: B: PDB Molecule: o-sialoglycoprotein endopeptidase/protein kinase;PDBTitle: structure of the methanococcus jannaschii kae1-bud32 fusion2 protein
50 c1f8uA_
not modelled
17.6
26
PDB header: hydrolase/toxinChain: A: PDB Molecule: acetylcholinesterase;PDBTitle: crystal structure of mutant e202q of human acetylcholinesterase2 complexed with green mamba venom peptide fasciculin-ii
51 d1f8ua_
not modelled
17.6
26
Fold: alpha/beta-HydrolasesSuperfamily: alpha/beta-HydrolasesFamily: Acetylcholinesterase-like52 d1s9aa_
not modelled
17.6
19
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase53 d2ozla1
not modelled
17.6
18
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: Branched-chain alpha-keto acid dehydrogenase PP module54 c3csvA_
not modelled
16.9
10
PDB header: transferaseChain: A: PDB Molecule: aminoglycoside phosphotransferase;PDBTitle: crystal structure of a putative aminoglycoside phosphotransferase2 (yp_614837.1) from silicibacter sp. tm1040 at 2.15 a resolution
55 d2pyqa1
not modelled
16.6
15
Fold: Jann4075-likeSuperfamily: Jann4075-likeFamily: Jann4075-like56 c2g5iB_
not modelled
16.3
17
PDB header: ligaseChain: B: PDB Molecule: aspartyl/glutamyl-trna(asn/gln) amidotransferasePDBTitle: structure of trna-dependent amidotransferase gatcab2 complexed with adp-alf4
57 d1ug0a_
not modelled
16.2
26
Fold: Surp module (SWAP domain)Superfamily: Surp module (SWAP domain)Family: Surp module (SWAP domain)58 c2w1jB_
not modelled
15.6
14
PDB header: transferaseChain: B: PDB Molecule: putative sortase;PDBTitle: crystal structure of sortase c-1 (srtc-1) from2 streptococcus pneumoniae
59 c3ecdC_
not modelled
15.3
13
PDB header: transferaseChain: C: PDB Molecule: serine hydroxymethyltransferase 2;PDBTitle: crystal structure of serine hydroxymethyltransferase from burkholderia2 pseudomallei
60 d3pccm_
not modelled
15.2
29
Fold: Prealbumin-likeSuperfamily: Aromatic compound dioxygenaseFamily: Aromatic compound dioxygenase61 c2q83A_
not modelled
15.1
18
PDB header: transferaseChain: A: PDB Molecule: ytaa protein;PDBTitle: crystal structure of ytaa (2635576) from bacillus subtilis at 2.50 a2 resolution
62 d2axtf1
not modelled
15.1
56
Fold: Single transmembrane helixSuperfamily: Cytochrome b559 subunitsFamily: Cytochrome b559 subunits63 c1w5cL_
not modelled
15.0
56
PDB header: photosynthesisChain: L: PDB Molecule: cytochrome b559 beta subunit;PDBTitle: photosystem ii from thermosynechococcus elongatus
64 d1xm8a_
not modelled
14.9
16
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Glyoxalase II (hydroxyacylglutathione hydrolase)65 d1q7ea_
not modelled
14.8
15
Fold: CoA-transferase family III (CaiB/BaiF)Superfamily: CoA-transferase family III (CaiB/BaiF)Family: CoA-transferase family III (CaiB/BaiF)66 c3izcs_
not modelled
14.4
35
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein rpl20 (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
67 c2x1cA_
not modelled
13.9
14
PDB header: transferaseChain: A: PDB Molecule: acyl-coenzymePDBTitle: the crystal structure of precursor acyl coenzyme2 a:isopenicillin n acyltransferase from penicillium3 chrysogenum
68 c2v1nA_
not modelled
13.9
19
PDB header: nuclear proteinChain: A: PDB Molecule: protein kin homolog;PDBTitle: solution structure of the region 51-160 of human kin172 reveals a winged helix fold
69 c2djcA_
not modelled
13.6
40
PDB header: cytokineChain: A: PDB Molecule: growth-blocking peptide;PDBTitle: solution structure of growth-blocking peptide of the2 tobacco cutworm, spodoptera litura
70 c3fc7B_
not modelled
13.6
20
PDB header: transferaseChain: B: PDB Molecule: htr-like protein;PDBTitle: the crystal structure of a domain of htr-like protein from haloarcula2 marismortui atcc 43049
71 c2dzrA_
not modelled
13.4
21
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-067, a gtf2i domain in human2 cdna
72 c3isyA_
not modelled
12.8
10
PDB header: protein bindingChain: A: PDB Molecule: intracellular proteinase inhibitor;PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
73 c3qd7X_
not modelled
12.5
26
PDB header: hydrolaseChain: X: PDB Molecule: uncharacterized protein ydal;PDBTitle: crystal structure of ydal, a stand-alone small muts-related protein2 from escherichia coli
74 d1ylxa1
not modelled
12.5
22
Fold: N domain of copper amine oxidase-likeSuperfamily: GK1464-likeFamily: GK1464-like75 c1irrA_
not modelled
12.5
40
PDB header: cytokineChain: A: PDB Molecule: paralytic peptide;PDBTitle: solution structure of paralytic peptide of the silkworm,2 bombyx mori
76 c1zawU_
not modelled
12.3
36
PDB header: structural proteinChain: U: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
77 c1zawW_
not modelled
12.3
36
PDB header: structural proteinChain: W: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
78 c1zawV_
not modelled
12.3
36
PDB header: structural proteinChain: V: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form a
79 c1zaxZ_
not modelled
12.1
36
PDB header: structural proteinChain: Z: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
80 d2d9ia1
not modelled
12.1
19
Fold: IF3-likeSuperfamily: SMR domain-likeFamily: Smr domain81 c3o0pA_
not modelled
11.9
8
PDB header: transferase , hydrolaseChain: A: PDB Molecule: sortase family protein;PDBTitle: pilus-related sortase c of group b streptococcus
82 c2hyiJ_
not modelled
11.8
44
PDB header: hydrolase/rna binding protein/rnaChain: J: PDB Molecule: protein casc3;PDBTitle: structure of the human exon junction complex with a trapped2 dead-box helicase bound to rna
83 c3i0oA_
not modelled
11.7
9
PDB header: transferaseChain: A: PDB Molecule: spectinomycin phosphotransferase;PDBTitle: crystal structure of spectinomycin phosphotransferase,2 aph(9)-ia, in complex with adp and spectinomcyin
84 d2pnwa1
not modelled
11.7
16
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: MLTA-like85 c3ex7I_
not modelled
11.6
40
PDB header: hydrolase/rna binding protein/rnaChain: I: PDB Molecule: protein casc3;PDBTitle: the crystal structure of ejc in its transition state
86 c1zaxY_
not modelled
11.5
37
PDB header: structural proteinChain: Y: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
87 c1zaxX_
not modelled
11.5
37
PDB header: structural proteinChain: X: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
88 c1zavY_
not modelled
11.5
37
PDB header: structural proteinChain: Y: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
89 c1zavX_
not modelled
11.5
37
PDB header: structural proteinChain: X: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
90 c2dzqA_
not modelled
11.5
17
PDB header: transcriptionChain: A: PDB Molecule: general transcription factor ii-i repeat domain-PDBTitle: solution structure of rsgi ruh-066, a gtf2i domain in human2 cdna
91 c1zavW_
not modelled
11.4
36
PDB header: structural proteinChain: W: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
92 c1zaxW_
not modelled
11.4
36
PDB header: structural proteinChain: W: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
93 c1zaxU_
not modelled
11.4
36
PDB header: structural proteinChain: U: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
94 c1zavU_
not modelled
11.4
36
PDB header: structural proteinChain: U: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
95 d1zavu1
not modelled
11.4
36
Fold: Ribosomal protein L7/12, oligomerisation (N-terminal) domainSuperfamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domainFamily: Ribosomal protein L7/12, oligomerisation (N-terminal) domain96 c1zavV_
not modelled
11.4
36
PDB header: structural proteinChain: V: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p21
97 c1zaxV_
not modelled
11.4
36
PDB header: structural proteinChain: V: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: ribosomal protein l10-l12(ntd) complex, space group p212121,2 form b
98 c1dd3C_
not modelled
11.2
36
PDB header: ribosomeChain: C: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima
99 c1dd3D_
not modelled
11.2
36
PDB header: ribosomeChain: D: PDB Molecule: 50s ribosomal protein l7/l12;PDBTitle: crystal structure of ribosomal protein l12 from thermotoga maritima