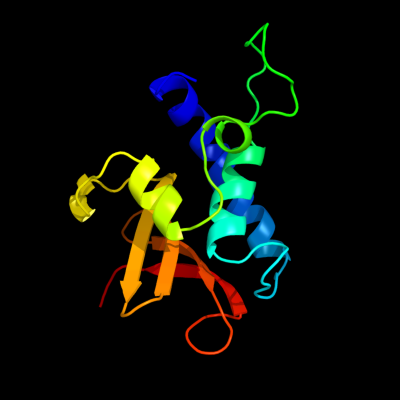
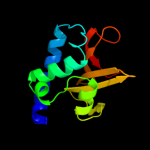
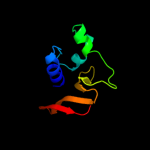
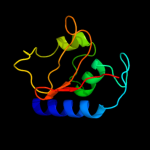
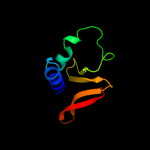
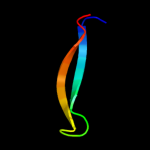
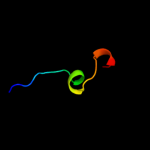
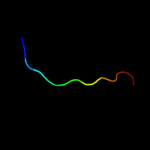
1 d2evra2
98.6
22
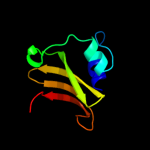
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: NlpC/P602 c2xivA_
98.6
24
PDB header: structural proteinChain: A: PDB Molecule: hypothetical invasion protein;PDBTitle: structure of rv1477, hypothetical invasion protein of2 mycobacterium tuberculosis
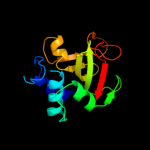
3 c3gt2A_
98.5
19
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium2 paratuberculosis antigen map1272c
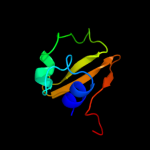
4 c3pbiA_
98.3
25
PDB header: hydrolaseChain: A: PDB Molecule: invasion protein;PDBTitle: structure of the peptidoglycan hydrolase ripb (rv1478) from2 mycobacterium tuberculosis at 1.6 resolution
5 c2fg0B_
98.3
24
PDB header: hydrolaseChain: B: PDB Molecule: cog0791: cell wall-associated hydrolases (invasion-PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (npun_r0659) from nostoc punctiforme pcc 73102 at 1.793 a resolution
6 c2k1gA_
98.3
15
PDB header: lipoproteinChain: A: PDB Molecule: lipoprotein spr;PDBTitle: solution nmr structure of lipoprotein spr from escherichia coli k12.2 northeast structural genomics target er541-37-162
7 c3i86A_
98.1
31
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of the p60 domain from m. avium subspecies2 paratuberculosis antigen map1204
8 c3npfB_
97.8
18
PDB header: hydrolaseChain: B: PDB Molecule: putative dipeptidyl-peptidase vi;PDBTitle: crystal structure of a putative dipeptidyl-peptidase vi (bacova_00612)2 from bacteroides ovatus at 1.72 a resolution
9 c3h41A_
96.4
26
PDB header: hydrolaseChain: A: PDB Molecule: nlp/p60 family protein;PDBTitle: crystal structure of a nlpc/p60 family protein (bce_2878) from2 bacillus cereus atcc 10987 at 1.79 a resolution
10 c3m1uB_
95.6
14
PDB header: hydrolaseChain: B: PDB Molecule: putative gamma-d-glutamyl-l-diamino acid endopeptidase;PDBTitle: crystal structure of a putative gamma-d-glutamyl-l-diamino acid2 endopeptidase (dvu_0896) from desulfovibrio vulgaris hildenborough at3 1.75 a resolution
11 c2kytA_
94.8
20
PDB header: hydrolaseChain: A: PDB Molecule: group xvi phospholipase a2;PDBTitle: solution struture of the h-rev107 n-terminal domain
12 c3kw0D_
92.1
22
PDB header: hydrolaseChain: D: PDB Molecule: cysteine peptidase;PDBTitle: crystal structure of cysteine peptidase (np_982244.1) from bacillus2 cereus atcc 10987 at 2.50 a resolution
13 d2if6a1
87.9
28
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: YiiX-like14 c2p1gA_
82.4
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative xylanase;PDBTitle: crystal structure of a putative xylanase from bacteroides fragilis
15 d2isba1
58.4
34
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: FumA C-terminal domain-likeFamily: FumA C-terminal domain-like16 c3kopB_
46.0
34
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein with a cyclophilin-like fold2 (yp_831253.1) from arthrobacter sp. fb24 at 1.90 a resolution
17 d1zcea1
34.0
21
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like18 d1t61c1
33.3
29
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Noncollagenous (NC1) domain of collagen IV19 d2iw0a1
29.4
26
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycoside hydrolase/deacetylaseFamily: NodB-like polysaccharide deacetylase20 d1t61a1
29.1
21
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Noncollagenous (NC1) domain of collagen IV21 d1o9ya_
not modelled
26.0
12
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)22 c2kncB_
not modelled
24.8
24
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
23 d1ofla_
not modelled
24.4
15
Fold: Single-stranded right-handed beta-helixSuperfamily: Pectin lyase-likeFamily: Chondroitinase B24 d1swxa_
not modelled
23.4
13
Fold: Glycolipid transfer protein, GLTPSuperfamily: Glycolipid transfer protein, GLTPFamily: Glycolipid transfer protein, GLTP25 c2iw0A_
not modelled
22.7
26
PDB header: hydrolaseChain: A: PDB Molecule: chitin deacetylase;PDBTitle: structure of the chitin deacetylase from the fungal2 pathogen colletotrichum lindemuthianum
26 d1li1c1
not modelled
22.5
21
Fold: C-type lectin-likeSuperfamily: C-type lectin-likeFamily: Noncollagenous (NC1) domain of collagen IV27 c2wbqA_
not modelled
22.0
40
PDB header: oxidoreductaseChain: A: PDB Molecule: l-arginine beta-hydroxylase;PDBTitle: crystal structure of vioc in complex with (2s,3s)-2 hydroxyarginine
28 d2ixma1
not modelled
21.5
25
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like29 d2gica1
not modelled
20.1
38
Fold: Rhabdovirus nucleoprotein-likeSuperfamily: Rhabdovirus nucleoprotein-likeFamily: Rhabdovirus nucleocapsid protein30 d1c0aa2
not modelled
19.7
57
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain31 c3emrA_
not modelled
19.6
18
PDB header: oxidoreductaseChain: A: PDB Molecule: ectd;PDBTitle: crystal structure analysis of the ectoine hydroxylase ectd from2 salibacillus salexigens
32 c2g62A_
not modelled
18.9
25
PDB header: hydrolase activatorChain: A: PDB Molecule: protein phosphatase 2a, regulatory subunit b' (pr 53);PDBTitle: crystal structure of human ptpa
33 d1o6aa_
not modelled
18.8
12
Fold: Surface presentation of antigens (SPOA)Superfamily: Surface presentation of antigens (SPOA)Family: Surface presentation of antigens (SPOA)34 d1ds1a_
not modelled
17.7
33
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Clavaminate synthase35 c3gipB_
not modelled
16.3
21
PDB header: hydrolaseChain: B: PDB Molecule: n-acyl-d-glutamate deacylase;PDBTitle: crystal structure of n-acyl-d-glutamate deacylase from2 bordetella bronchiseptica complexed with zinc, acetate and3 formate ions.
36 c1dbgA_
not modelled
15.4
15
PDB header: lyaseChain: A: PDB Molecule: chondroitinase b;PDBTitle: crystal structure of chondroitinase b
37 d2ar1a1
not modelled
15.2
13
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like38 d1l0wa2
not modelled
15.2
36
Fold: DCoH-likeSuperfamily: GAD domain-likeFamily: GAD domain39 c1dfwA_
not modelled
15.1
100
PDB header: immune systemChain: A: PDB Molecule: lung surfactant protein b;PDBTitle: conformational mapping of the n-terminal segment of2 surfactant protein b in lipid using 13c-enhanced fourier3 transform infrared spectroscopy (ftir)
40 d1hq0a_
not modelled
14.8
19
Fold: CNF1/YfiH-like putative cysteine hydrolasesSuperfamily: CNF1/YfiH-like putative cysteine hydrolasesFamily: Type 1 cytotoxic necrotizing factor, catalytic domain41 d2gtta1
not modelled
14.6
56
Fold: Rhabdovirus nucleoprotein-likeSuperfamily: Rhabdovirus nucleoprotein-likeFamily: Rhabdovirus nucleocapsid protein42 d1vk3a3
not modelled
14.3
30
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like43 c2ysxA_
not modelled
13.5
9
PDB header: signaling proteinChain: A: PDB Molecule: signaling inositol polyphosphate phosphatasePDBTitle: solution structure of the human ship sh2 domain
44 d1leha2
not modelled
13.4
25
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases45 d2adza1
not modelled
13.3
20
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)46 c2fvmA_
not modelled
13.2
14
PDB header: hydrolaseChain: A: PDB Molecule: dihydropyrimidinase;PDBTitle: crystal structure of dihydropyrimidinase from saccharomyces kluyveri2 in complex with the reaction product n-carbamyl-beta-alanine
47 c1rjqA_
not modelled
13.0
21
PDB header: hydrolaseChain: A: PDB Molecule: d-aminoacylase;PDBTitle: the crystal structure of the d-aminoacylase mutant d366a
48 c2rdsA_
not modelled
13.0
36
PDB header: oxidoreductaseChain: A: PDB Molecule: 1-deoxypentalenic acid 11-beta hydroxylase; fe(ii)/alpha-PDBTitle: crystal structure of ptlh with fe/oxalylglycine and ent-1-2 deoxypentalenic acid bound
49 c3nnlB_
not modelled
13.0
40
PDB header: biosynthetic proteinChain: B: PDB Molecule: cura;PDBTitle: halogenase domain from cura module (crystal form iii)
50 c2og5A_
not modelled
13.0
33
PDB header: oxidoreductaseChain: A: PDB Molecule: putative oxygenase;PDBTitle: crystal structure of asparagine oxygenase (asno)
51 d2ixna1
not modelled
12.9
16
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like52 d1hcza1
not modelled
12.4
64
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Cytochrome f, large domainFamily: Cytochrome f, large domain53 d1pvda1
not modelled
12.4
30
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain54 c1t60W_
not modelled
12.3
25
PDB header: structural proteinChain: W: PDB Molecule: type iv collagen;PDBTitle: crystal structure of type iv collagen nc1 domain from2 bovine lens capsule
55 d2z1ea2
not modelled
12.2
38
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like56 c3lv4B_
not modelled
12.0
26
PDB header: hydrolaseChain: B: PDB Molecule: glycoside hydrolase yxia;PDBTitle: crystal structure of the glycoside hydrolase, family 43 yxia2 protein from bacillus licheniformis. northeast structural3 genomics consortium target bir14.
57 d1jz8a3
not modelled
11.9
27
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: beta-Galactosidase/glucuronidase, N-terminal domain58 c3mt1B_
not modelled
11.8
14
PDB header: lyaseChain: B: PDB Molecule: putative carboxynorspermidine decarboxylase protein;PDBTitle: crystal structure of putative carboxynorspermidine decarboxylase2 protein from sinorhizobium meliloti
59 d2a1xa1
not modelled
11.8
36
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: PhyH-like60 c1t60R_
not modelled
11.4
25
PDB header: structural proteinChain: R: PDB Molecule: type iv collagen;PDBTitle: crystal structure of type iv collagen nc1 domain from2 bovine lens capsule
61 d1fi4a2
not modelled
11.2
15
Fold: Ferredoxin-likeSuperfamily: GHMP Kinase, C-terminal domainFamily: Mevalonate 5-diphosphate decarboxylase62 c1lehB_
not modelled
11.0
36
PDB header: oxidoreductaseChain: B: PDB Molecule: leucine dehydrogenase;PDBTitle: leucine dehydrogenase from bacillus sphaericus
63 d1iufa1
not modelled
10.9
26
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Centromere-binding64 d1zpda1
not modelled
10.6
30
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain65 c1fi4A_
not modelled
10.5
15
PDB header: lyaseChain: A: PDB Molecule: mevalonate 5-diphosphate decarboxylase;PDBTitle: the x-ray crystal structure of mevalonate 5-diphosphate decarboxylase2 at 2.3 angstrom resolution.
66 d1ovma1
not modelled
10.5
30
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Pyruvate oxidase and decarboxylase, middle domain67 d2zoda2
not modelled
10.3
25
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like68 d2gbsa1
not modelled
10.2
12
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: Atu2648/PH1033-like69 d1c1da2
not modelled
9.8
29
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Aminoacid dehydrogenases70 c2kafA_
not modelled
9.8
28
PDB header: viral protein, rna binding proteinChain: A: PDB Molecule: non-structural protein 3;PDBTitle: solution structure of the sars-unique domain-c from the2 nonstructural protein 3 (nsp3) of the severe acute3 respiratory syndrome coronavirus
71 d1o5ua_
not modelled
9.8
33
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Hypothetical protein TM111272 c2opwA_
not modelled
9.7
18
PDB header: oxidoreductaseChain: A: PDB Molecule: phyhd1 protein;PDBTitle: crystal structure of human phytanoyl-coa dioxygenase phyhd1 (apo)
73 d2j0pa1
not modelled
9.5
38
Fold: Heme iron utilization protein-likeSuperfamily: Heme iron utilization protein-likeFamily: HemS/ChuS-like74 d2coha2
not modelled
9.2
12
Fold: Double-stranded beta-helixSuperfamily: cAMP-binding domain-likeFamily: cAMP-binding domain75 c3hqjA_
not modelled
9.2
40
PDB header: transferaseChain: A: PDB Molecule: holo-[acyl-carrier-protein] synthase;PDBTitle: structure-function analysis of mycobacterium tuberculosis2 acyl carrier protein synthase (acps).
76 d1gxha_
not modelled
9.1
36
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins77 d1k3xa2
not modelled
8.9
18
Fold: N-terminal domain of MutM-like DNA repair proteinsSuperfamily: N-terminal domain of MutM-like DNA repair proteinsFamily: N-terminal domain of MutM-like DNA repair proteins78 c2y9xG_
not modelled
8.9
44
PDB header: oxidoreductaseChain: G: PDB Molecule: lectin-like fold protein;PDBTitle: crystal structure of ppo3, a tyrosinase from agaricus bisporus, in2 deoxy-form that contains additional unknown lectin-like subunit,3 with inhibitor tropolone
79 c3k2zA_
not modelled
8.8
14
PDB header: hydrolaseChain: A: PDB Molecule: lexa repressor;PDBTitle: crystal structure of a lexa protein from thermotoga maritima
80 d2es7a1
not modelled
8.7
24
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: HyaE-like81 c3f0nB_
not modelled
8.7
20
PDB header: lyaseChain: B: PDB Molecule: mevalonate pyrophosphate decarboxylase;PDBTitle: mus musculus mevalonate pyrophosphate decarboxylase
82 c2uz8A_
not modelled
8.6
18
PDB header: rna-binding proteinChain: A: PDB Molecule: eukaryotic translation elongation factor 1PDBTitle: the crystal structure of p18, human translation elongation2 factor 1 epsilon 1
83 c3mazA_
not modelled
8.5
14
PDB header: signaling proteinChain: A: PDB Molecule: signal-transducing adaptor protein 1;PDBTitle: crystal structure of the human brdg1/stap-1 sh2 domain in complex with2 the ntal ptyr136 peptide
84 d2io8a2
not modelled
8.2
19
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: CHAP domain85 c1zzpA_
not modelled
8.2
18
PDB header: transferaseChain: A: PDB Molecule: proto-oncogene tyrosine-protein kinase abl1;PDBTitle: solution structure of the f-actin binding domain of bcr-2 abl/c-abl
86 d1ayia_
not modelled
8.1
27
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins87 c2hnfA_
not modelled
8.1
24
PDB header: viral proteinChain: A: PDB Molecule: repressor protein ci101-229dm-k192a;PDBTitle: structure of a hyper-cleavable monomeric fragment of phage2 lambda repressor containing the cleavage site region
88 d2ixoa1
not modelled
8.0
6
Fold: PTPA-likeSuperfamily: PTPA-likeFamily: PTPA-like89 d2vlqa1
not modelled
7.6
36
Fold: Acyl carrier protein-likeSuperfamily: Colicin E immunity proteinsFamily: Colicin E immunity proteins90 c2no8A_
not modelled
7.6
36
PDB header: immune systemChain: A: PDB Molecule: colicin-e2 immunity protein;PDBTitle: nmr structure analysis of the colicin immuntiy protein im2
91 c2jx0A_
not modelled
7.5
24
PDB header: cell adhesion, signaling proteinChain: A: PDB Molecule: arf gtpase-activating protein git1;PDBTitle: the paxillin-binding domain (pbd) of g protein coupled2 receptor (gpcr)-kinase (grk) interacting protein 1 (git1)
92 d1c3ga1
not modelled
7.5
13
Fold: HSP40/DnaJ peptide-binding domainSuperfamily: HSP40/DnaJ peptide-binding domainFamily: HSP40/DnaJ peptide-binding domain93 d2pyta1
not modelled
7.5
16
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: EutQ-like94 d3c9ua2
not modelled
7.4
50
Fold: PurM C-terminal domain-likeSuperfamily: PurM C-terminal domain-likeFamily: PurM C-terminal domain-like95 c3qrxB_
not modelled
7.3
67
PDB header: metal binding protein/toxinChain: B: PDB Molecule: melittin;PDBTitle: chlamydomonas reinhardtii centrin bound to melittin
96 d1uv4a1
not modelled
7.3
23
Fold: 5-bladed beta-propellerSuperfamily: Arabinanase/levansucrase/invertaseFamily: alpha-L-arabinanase-like97 d2c9wa2
not modelled
7.1
11
Fold: SH2-likeSuperfamily: SH2 domainFamily: SH2 domain98 d1jr7a_
not modelled
7.1
13
Fold: Double-stranded beta-helixSuperfamily: Clavaminate synthase-likeFamily: Gab protein (hypothetical protein YgaT)99 c2jemB_
not modelled
7.0
26
PDB header: hydrolaseChain: B: PDB Molecule: endo-beta-1,4-glucanase;PDBTitle: native family 12 xyloglucanase from bacillus licheniformis