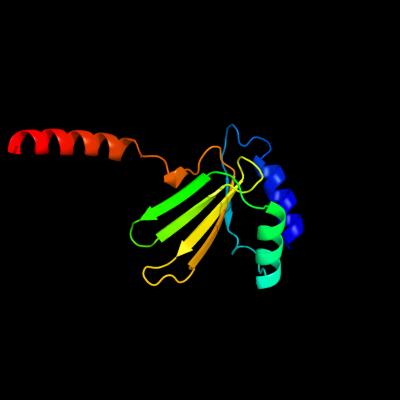
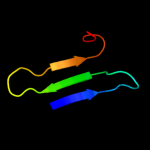
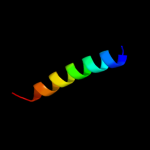
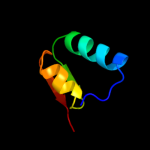
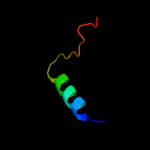
1 c3sb1B_
100.0
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: hydrogenase expression protein;PDBTitle: hydrogenase expression protein huph from thiobacillus denitrificans2 atcc 25259
2 c1vp7D_
36.1
13
PDB header: hydrolaseChain: D: PDB Molecule: exodeoxyribonuclease vii small subunit;PDBTitle: crystal structure of exodeoxyribonuclease vii small subunit2 (np_881400.1) from bordetella pertussis at 2.40 a resolution
3 d1vp7a_
29.7
13
Fold: Spectrin repeat-likeSuperfamily: XseB-likeFamily: XseB-like4 d1vp7b_
25.9
13
Fold: Spectrin repeat-likeSuperfamily: XseB-likeFamily: XseB-like5 c2ka6B_
24.7
27
PDB header: transcription regulatorChain: B: PDB Molecule: signal transducer and activator of transcriptionPDBTitle: nmr structure of the cbp-taz2/stat1-tad complex
6 c2w56B_
24.1
13
PDB header: unknown functionChain: B: PDB Molecule: vc0508;PDBTitle: structure of the hypothetical protein vc0508 from vibrio cholerae2 vsp-ii pathogenicity island
7 d1vhib_
20.0
43
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain8 c3gkuB_
19.7
11
PDB header: rna binding proteinChain: B: PDB Molecule: probable rna-binding protein;PDBTitle: crystal structure of a probable rna-binding protein from clostridium2 symbiosum atcc 14940
9 d1p3wa_
18.5
13
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like10 d1b3ta_
16.4
50
Fold: Ferredoxin-likeSuperfamily: Viral DNA-binding domainFamily: Viral DNA-binding domain11 c3h8dC_
14.6
15
PDB header: motor protein/signaling proteinChain: C: PDB Molecule: myosin-vi;PDBTitle: crystal structure of myosin vi in complex with dab2 peptide
12 c3lvmB_
14.0
13
PDB header: transferaseChain: B: PDB Molecule: cysteine desulfurase;PDBTitle: crystal structure of e.coli iscs
13 c3cvoA_
13.3
15
PDB header: transferaseChain: A: PDB Molecule: methyltransferase-like protein of unknown function;PDBTitle: crystal structure of a methyltransferase-like protein (spo2022) from2 silicibacter pomeroyi dss-3 at 1.80 a resolution
14 c3d5jB_
12.5
15
PDB header: oxidoreductaseChain: B: PDB Molecule: glutaredoxin-2, mitochondrial;PDBTitle: structure of yeast grx2-c30s mutant with glutathionyl mixed2 disulfide
15 d2f2ab1
12.3
13
Fold: GatB/YqeY motifSuperfamily: GatB/YqeY motifFamily: GatB/GatE C-terminal domain-like16 c3if8B_
12.3
15
PDB header: cell cycleChain: B: PDB Molecule: protein zwilch homolog;PDBTitle: crystal structure of zwilch, a member of the rzz kinetochore complex
17 d1gyva_
12.1
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like18 c3n6qF_
12.0
17
PDB header: oxidoreductaseChain: F: PDB Molecule: yghz aldo-keto reductase;PDBTitle: crystal structure of yghz from e. coli
19 d1gywb_
11.9
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like20 d2ns0a1
11.5
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: RHA1 ro06458-like21 d1th0a_
not modelled
10.6
18
Fold: Cysteine proteinasesSuperfamily: Cysteine proteinasesFamily: Adenain-like22 c2qf7A_
not modelled
10.5
17
PDB header: ligaseChain: A: PDB Molecule: pyruvate carboxylase protein;PDBTitle: crystal structure of a complete multifunctional pyruvate carboxylase2 from rhizobium etli
23 c3ipzA_
not modelled
10.2
11
PDB header: electron transport, oxidoreductaseChain: A: PDB Molecule: monothiol glutaredoxin-s14, chloroplastic;PDBTitle: crystal structure of arabidopsis monothiol glutaredoxin atgrxcp
24 c2wj8N_
not modelled
9.9
13
PDB header: rna binding protein/rnaChain: N: PDB Molecule: nucleoprotein;PDBTitle: respiratory syncitial virus ribonucleoprotein
25 d2bz2a1
not modelled
9.7
2
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD26 c1mszA_
not modelled
9.0
19
PDB header: dna binding proteinChain: A: PDB Molecule: dna-binding protein smubp-2;PDBTitle: solution structure of the r3h domain from human smubp-2
27 d1msza_
not modelled
9.0
19
Fold: IF3-likeSuperfamily: R3H domainFamily: R3H domain28 c2kwtA_
not modelled
8.8
29
PDB header: viral proteinChain: A: PDB Molecule: protease ns2-3;PDBTitle: solution structure of ns2 [27-59]
29 c2f3jA_
not modelled
8.6
11
PDB header: transport proteinChain: A: PDB Molecule: rna and export factor binding protein 2;PDBTitle: the solution structure of the ref2-i mrna export factor2 (residues 1-155).
30 d2ppqa1
not modelled
8.2
10
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: APH phosphotransferases31 c3exnA_
not modelled
8.2
27
PDB header: transferaseChain: A: PDB Molecule: probable acetyltransferase;PDBTitle: crystal structure of acetyltransferase from thermus thermophilus hb8
32 c3c1sA_
not modelled
8.1
15
PDB header: oxidoreductaseChain: A: PDB Molecule: glutaredoxin-1;PDBTitle: crystal structure of grx1 in glutathionylated form
33 d1qk1a2
not modelled
8.0
9
Fold: Glutamine synthetase/guanido kinaseSuperfamily: Glutamine synthetase/guanido kinaseFamily: Guanido kinase catalytic domain34 d2dwya1
not modelled
7.8
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like35 d1na8a_
not modelled
6.9
9
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Clathrin adaptor appendage domainFamily: gamma-adaptin C-terminal appendage domain-like36 c2huuA_
not modelled
6.8
22
PDB header: transferaseChain: A: PDB Molecule: alanine glyoxylate aminotransferase;PDBTitle: crystal structure of aedes aegypti alanine glyoxylate2 aminotransferase in complex with alanine
37 d1j9ba_
not modelled
6.8
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: ArsC-like38 c2e9gA_
not modelled
6.7
14
PDB header: protein bindingChain: A: PDB Molecule: ap-1 complex subunit gamma-2;PDBTitle: solution structure of the alpha adaptinc2 domain from human2 adapter-related protein complex 1 gamma 2 subunit
39 c2ee6A_
not modelled
6.4
17
PDB header: structural proteinChain: A: PDB Molecule: filamin-b;PDBTitle: solution structure of the 21th filamin domain from human2 filamin-b
40 d1qmra_
not modelled
6.3
14
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like41 d2bk0a1
not modelled
6.2
12
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Pathogenesis-related protein 10 (PR10)-like42 d1t07a_
not modelled
6.1
19
Fold: YggX-likeSuperfamily: YggX-likeFamily: YggX-like43 c3e4fB_
not modelled
5.9
17
PDB header: transferaseChain: B: PDB Molecule: aminoglycoside n3-acetyltransferase;PDBTitle: crystal structure of ba2930- a putative aminoglycoside n3-2 acetyltransferase from bacillus anthracis
44 d1bwwa_
not modelled
5.8
12
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)45 c2hbpA_
not modelled
5.8
17
PDB header: endocytosis, protein bindingChain: A: PDB Molecule: cytoskeleton assembly control protein sla1;PDBTitle: solution structure of sla1 homology domain 1
46 d2b79a1
not modelled
5.7
11
Fold: TBP-likeSuperfamily: Bet v1-likeFamily: Smu440-like47 d1y7ta2
not modelled
5.7
11
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain48 d1c7na_
not modelled
5.7
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like49 c2hqyB_
not modelled
5.6
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: conserved hypothetical protein;PDBTitle: crystal structure of conserved protein of unknown function from2 bacteroides thetaiotaomicron vpi-5482
50 c3rv2B_
not modelled
5.6
41
PDB header: transferaseChain: B: PDB Molecule: s-adenosylmethionine synthase;PDBTitle: crystal structure of s-adenosylmethionine synthetase from2 mycobacterium marinum
51 d1hy7a_
not modelled
5.5
24
Fold: Zincin-likeSuperfamily: Metalloproteases ("zincins"), catalytic domainFamily: Matrix metalloproteases, catalytic domain52 d2bp3a1
not modelled
5.5
20
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Filamin repeat (rod domain)53 d3cmol1
not modelled
5.4
13
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)54 c1y03A_
not modelled
5.2
43
PDB header: antifreeze proteinChain: A: PDB Molecule: antifreeze peptide ss-3;PDBTitle: solution structure of a recombinant type i sculpin2 antifreeze protein
55 c1y04A_
not modelled
5.2
43
PDB header: antifreeze proteinChain: A: PDB Molecule: antifreeze peptide ss-3;PDBTitle: solution structure of a recombinant type i sculpin2 antifreeze protein
56 d2ad9a1
not modelled
5.1
21
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD