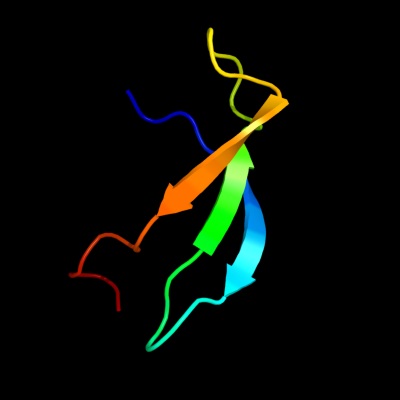
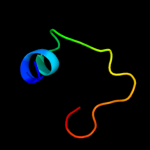
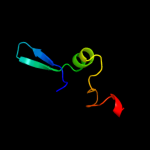
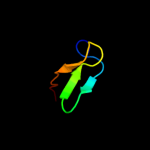
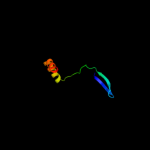
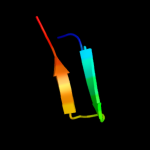
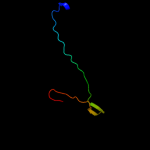
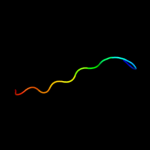
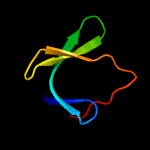
1 d1ah9a_
61.4
26
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like2 c3i4oA_
57.8
17
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-1;PDBTitle: crystal structure of translation initiation factor 1 from2 mycobacterium tuberculosis
3 d1m9sa4
51.1
19
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: GW domain4 d1tt2a_
49.4
12
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease5 c3bdlA_
48.0
25
PDB header: hydrolaseChain: A: PDB Molecule: staphylococcal nuclease domain-containingPDBTitle: crystal structure of a truncated human tudor-sn
6 d2ix0a3
35.4
11
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like7 d2ijra1
32.7
27
Fold: Api92-likeSuperfamily: Api92-likeFamily: Api92-like8 d1snoa_
29.7
12
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease9 c3etcB_
29.6
32
PDB header: ligaseChain: B: PDB Molecule: amp-binding protein;PDBTitle: 2.1 a structure of acyl-adenylate synthetase from methanosarcina2 acetivorans containing a link between lys256 and cys298
10 d1khca_
26.7
24
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain11 c2yyjA_
24.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: 4-hydroxyphenylacetate-3-hydroxylase;PDBTitle: crystal structure of the oxygenase component (hpab) of 4-2 hydroxyphenylacetate 3-monooxygenase complexed with fad and 4-3 hydroxyphenylacetate
12 c3pzvB_
24.3
22
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase;PDBTitle: c2 crystal form of the endo-1,4-beta-glucanase from bacillus subtilis2 168
13 d1vlba3
23.2
22
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like14 d1dgja3
22.4
30
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like15 c3f8tA_
22.3
13
PDB header: hydrolaseChain: A: PDB Molecule: predicted atpase involved in replication control,PDBTitle: crystal structure analysis of a full-length mcm homolog2 from methanopyrus kandleri
16 c3izcG_
22.0
25
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein rpl6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
17 d1nbwa2
21.9
27
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ATPase domain of dehydratase reactivase alpha subunit18 c2l66B_
21.8
23
PDB header: transcription regulatorChain: B: PDB Molecule: transcriptional regulator, abrb family;PDBTitle: the dna-recognition fold of sso7c4 suggests a new member of spovt-abrb2 superfamily from archaea.
19 c2dzcA_
21.0
15
PDB header: ligaseChain: A: PDB Molecule: biotin--[acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from pyrococcus2 horikoshii, mutation r48a
20 c3llrA_
20.4
35
PDB header: transferaseChain: A: PDB Molecule: dna (cytosine-5)-methyltransferase 3a;PDBTitle: crystal structure of the pwwp domain of human dna (cytosine-5-)-2 methyltransferase 3 alpha
21 d1hr0w_
not modelled
20.2
24
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like22 d1qupa2
not modelled
18.8
22
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain23 d1n62b1
not modelled
18.7
13
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like24 d1fvia1
not modelled
18.3
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain25 c2wctC_
not modelled
18.0
25
PDB header: rna-binding proteinChain: C: PDB Molecule: non-structural protein 3;PDBTitle: human sars coronavirus unique domain (triclinic form)
26 d1m9sa2
not modelled
17.0
13
Fold: SH3-like barrelSuperfamily: Prokaryotic SH3-related domainFamily: GW domain27 d1rkna_
not modelled
16.3
12
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease28 d2k0bx1
not modelled
15.7
31
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain29 d1ng2a2
not modelled
13.2
14
Fold: SH3-like barrelSuperfamily: SH3-domainFamily: SH3-domain30 d1ffvb1
not modelled
12.8
13
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like31 c2wj7D_
not modelled
12.8
14
PDB header: chaperoneChain: D: PDB Molecule: alpha-crystallin b chain;PDBTitle: human alphab crystallin
32 d1kj1a_
not modelled
12.6
27
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins33 c2b9kA_
not modelled
12.3
46
PDB header: antibioticChain: A: PDB Molecule: antimicrobial peptide lci;PDBTitle: solution structure of lci, an amp from bacillus subtilis
34 d2essa1
not modelled
12.0
7
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: Acyl-ACP thioesterase-like35 d1t3qb1
not modelled
12.0
26
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like36 d2hlja1
not modelled
11.9
9
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: 4HBT-like37 d2soba_
not modelled
11.7
10
Fold: OB-foldSuperfamily: Staphylococcal nucleaseFamily: Staphylococcal nuclease38 c2crlA_
not modelled
11.5
7
PDB header: chaperoneChain: A: PDB Molecule: copper chaperone for superoxide dismutase;PDBTitle: the apo form of hma domain of copper chaperone for2 superoxide dismutase
39 d1tdja3
not modelled
11.2
31
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Allosteric threonine deaminase C-terminal domain40 c3r0eD_
not modelled
11.1
15
PDB header: sugar binding proteinChain: D: PDB Molecule: lectin;PDBTitle: structure of remusatia vivipara lectin
41 c3mezB_
not modelled
10.6
36
PDB header: sugar binding proteinChain: B: PDB Molecule: mannose-specific lectin 3 chain 2;PDBTitle: x-ray structural analysis of a mannose specific lectin from dutch2 crocus (crocus vernus)
42 c2oi3A_
not modelled
10.5
15
PDB header: transferaseChain: A: PDB Molecule: tyrosine-protein kinase hck;PDBTitle: nmr structure analysis of the hematopoetic cell kinase sh32 domain complexed with an artificial high affinity ligand3 (pd1)
43 d1cc8a_
not modelled
10.3
12
Fold: Ferredoxin-likeSuperfamily: HMA, heavy metal-associated domainFamily: HMA, heavy metal-associated domain44 d1g2913
not modelled
10.2
17
Fold: OB-foldSuperfamily: MOP-likeFamily: ABC-transporter additional domain45 c2ewnA_
not modelled
10.1
12
PDB header: ligase, transcriptionChain: A: PDB Molecule: bira bifunctional protein;PDBTitle: ecoli biotin repressor with co-repressor analog
46 c3ipfA_
not modelled
9.6
19
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the q251q8_deshy protein from desulfitobacterium2 hafniense. northeast structural genomics consortium target dhr8c.
47 d1bwud_
not modelled
9.5
21
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins48 d1hnga1
not modelled
9.4
40
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)49 c2wgnB_
not modelled
9.4
25
PDB header: hydrolase inhibitorChain: B: PDB Molecule: inhibitor of cysteine peptidase compnd 3;PDBTitle: pseudomonas aeruginosa icp
50 d7a3ha_
not modelled
9.2
17
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases51 c2kctA_
not modelled
9.1
21
PDB header: chaperoneChain: A: PDB Molecule: cytochrome c-type biogenesis protein ccme;PDBTitle: solution nmr structure of the ob-fold domain of heme2 chaperone ccme from desulfovibrio vulgaris. northeast3 structural genomics target dvr115g.
52 d1b2pa_
not modelled
9.1
12
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins53 d1vqoq1
not modelled
8.6
7
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal proteins L24p and L21e54 c2dpfB_
not modelled
8.6
14
PDB header: plant proteinChain: B: PDB Molecule: curculin;PDBTitle: crystal structure of curculin1 homodimer
55 d1v25a_
not modelled
8.5
12
Fold: Acetyl-CoA synthetase-likeSuperfamily: Acetyl-CoA synthetase-likeFamily: Acetyl-CoA synthetase-like56 c2zkrq_
not modelled
8.4
30
PDB header: ribosomal protein/rnaChain: Q: PDB Molecule: rna expansion segment es31 part ii;PDBTitle: structure of a mammalian ribosomal 60s subunit within an2 80s complex obtained by docking homology models of the rna3 and proteins into an 8.7 a cryo-em map
57 c4a1dE_
not modelled
8.4
38
PDB header: ribosomeChain: E: PDB Molecule: rpl6;PDBTitle: t.thermophila 60s ribosomal subunit in complex with initiation2 factor 6. this file contains 26s rrna and proteins of3 molecule 4.
58 d1kj1d_
not modelled
8.2
29
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins59 c1s1iQ_
not modelled
8.2
22
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l21-a;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1i,4 contains 60s subunit. the 40s ribosomal subunit is in file5 1s1h.
60 c3r0eC_
not modelled
8.2
36
PDB header: sugar binding proteinChain: C: PDB Molecule: lectin;PDBTitle: structure of remusatia vivipara lectin
61 c2eayB_
not modelled
8.2
20
PDB header: ligaseChain: B: PDB Molecule: biotin [acetyl-coa-carboxylase] ligase;PDBTitle: crystal structure of biotin protein ligase from aquifex2 aeolicus
62 c1w3gA_
not modelled
8.0
16
PDB header: toxin/lectinChain: A: PDB Molecule: hemolytic lectin from laetiporus sulphureus;PDBTitle: hemolytic lectin from the mushroom laetiporus sulphureus2 complexed with two n-acetyllactosamine molecules.
63 d1jrob1
not modelled
7.9
13
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like64 c3iz5U_
not modelled
7.9
26
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein l21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
65 c2cghB_
not modelled
7.8
8
PDB header: ligaseChain: B: PDB Molecule: biotin ligase;PDBTitle: crystal structure of biotin ligase from mycobacterium2 tuberculosis
66 c4a1aP_
not modelled
7.7
13
PDB header: ribosomeChain: P: PDB Molecule: 60s ribosomal protein l21;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
67 d1npla_
not modelled
7.7
20
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins68 d1kafa_
not modelled
7.7
29
Fold: MotA C-terminal domain-likeSuperfamily: DNA-binding C-terminal domain of the transcription factor MotAFamily: DNA-binding C-terminal domain of the transcription factor MotA69 d1khda2
not modelled
7.6
19
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain70 c3elqA_
not modelled
7.6
18
PDB header: transferaseChain: A: PDB Molecule: arylsulfate sulfotransferase;PDBTitle: crystal structure of a bacterial arylsulfate2 sulfotransferase
71 d1hnfa1
not modelled
7.6
14
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)72 c3m86B_
not modelled
7.4
31
PDB header: protein bindingChain: B: PDB Molecule: amoebiasin-2;PDBTitle: crystal structure of the cysteine protease inhibitor, ehicp2, from2 entamoeba histolytica
73 d1bwua_
not modelled
7.4
21
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins74 d2joya1
not modelled
7.4
13
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L14e75 c2w54F_
not modelled
7.4
13
PDB header: oxidoreductaseChain: F: PDB Molecule: xanthine dehydrogenase;PDBTitle: crystal structure of xanthine dehydrogenase from2 rhodobacter capsulatus in complex with bound inhibitor3 pterin-6-aldehyde
76 c3izcU_
not modelled
7.4
22
PDB header: ribosomeChain: U: PDB Molecule: 60s ribosomal protein rpl21 (l21e);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
77 d1d7qa_
not modelled
7.3
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like78 d2elca2
not modelled
7.3
19
Fold: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainSuperfamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domainFamily: Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain79 c2imzA_
not modelled
7.2
7
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease pi-mtui;PDBTitle: crystal structure of mtu reca intein splicing domain
80 c3ij3A_
not modelled
7.1
25
PDB header: hydrolaseChain: A: PDB Molecule: cytosol aminopeptidase;PDBTitle: 1.8 angstrom resolution crystal structure of cytosol aminopeptidase2 from coxiella burnetii
81 d1jpca_
not modelled
7.1
20
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins82 c2oqkA_
not modelled
7.1
19
PDB header: translationChain: A: PDB Molecule: putative translation initiation factor eif-1a;PDBTitle: crystal structure of putative cryptosporidium parvum translation2 initiation factor eif-1a
83 c2dbkA_
not modelled
7.0
12
PDB header: signaling proteinChain: A: PDB Molecule: crk-like protein;PDBTitle: solution structures of the sh3 domain of human crk-like2 protein
84 d1oxxk1
not modelled
7.0
22
Fold: OB-foldSuperfamily: MOP-likeFamily: ABC-transporter additional domain85 c2klrA_
not modelled
6.9
14
PDB header: chaperoneChain: A: PDB Molecule: alpha-crystallin b chain;PDBTitle: solid-state nmr structure of the alpha-crystallin domain in alphab-2 crystallin oligomers
86 d1l4zb_
not modelled
6.9
26
Fold: beta-Grasp (ubiquitin-like)Superfamily: Staphylokinase/streptokinaseFamily: Staphylokinase/streptokinase87 c2k5hA_
not modelled
6.8
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein;PDBTitle: solution nmr structure of protein encoded by mth693 from2 methanobacterium thermoautotrophicum: northeast structural3 genomics consortium target tt824a
88 d2zjrm1
not modelled
6.8
27
Fold: SH3-like barrelSuperfamily: Translation proteins SH3-like domainFamily: Ribosomal protein L1989 c1u8vA_
not modelled
6.7
20
PDB header: lyase, isomeraseChain: A: PDB Molecule: gamma-aminobutyrate metabolismPDBTitle: crystal structure of 4-hydroxybutyryl-coa dehydratase from2 clostridium aminobutyricum: radical catalysis involving a3 [4fe-4s] cluster and flavin
90 d2exda1
not modelled
6.7
20
Fold: OB-foldSuperfamily: NfeD domain-likeFamily: NfeD domain-like91 c1qupA_
not modelled
6.7
22
PDB header: chaperoneChain: A: PDB Molecule: superoxide dismutase 1 copper chaperone;PDBTitle: crystal structure of the copper chaperone for superoxide2 dismutase
92 c3iuwA_
not modelled
6.7
36
PDB header: rna binding proteinChain: A: PDB Molecule: activating signal cointegrator;PDBTitle: crystal structure of activating signal cointegrator (np_814290.1) from2 enterococcus faecalis v583 at 1.58 a resolution
93 c3iz5G_
not modelled
6.6
31
PDB header: ribosomeChain: G: PDB Molecule: 60s ribosomal protein l6 (l6e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
94 d1jt8a_
not modelled
6.5
16
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like95 c3h8gC_
not modelled
6.4
25
PDB header: hydrolaseChain: C: PDB Molecule: cytosol aminopeptidase;PDBTitle: bestatin complex structure of leucine aminopeptidase from pseudomonas2 putida
96 c2zs6B_
not modelled
6.4
22
PDB header: toxinChain: B: PDB Molecule: hemagglutinin components ha3;PDBTitle: ha3 subcomponent of botulinum type c progenitor toxin
97 c2e4mC_
not modelled
6.3
44
PDB header: toxinChain: C: PDB Molecule: ha-17;PDBTitle: crystal structure of hemagglutinin subcomponent complex (ha-2 33/ha-17) from clostridium botulinum serotype d strain 4947
98 d1v97a3
not modelled
6.3
13
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like99 d3b9jc1
not modelled
6.1
13
Fold: alpha/beta-HammerheadSuperfamily: CO dehydrogenase molybdoprotein N-domain-likeFamily: CO dehydrogenase molybdoprotein N-domain-like