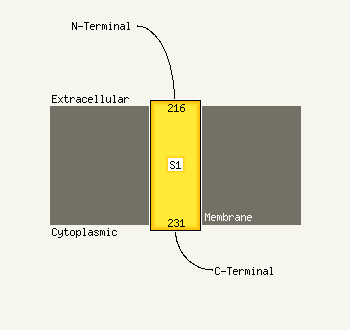
1 c2k5eA_
28.3
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of putative uncharacterized protein2 gsu1278 from methanocaldococcus jannaschii, northeast3 structural genomics consortium (nesg) target gsr195
2 d2uube1
20.1
19
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Translational machinery components3 c2pr1B_
18.0
50
PDB header: transferaseChain: B: PDB Molecule: uncharacterized n-acetyltransferase ylbp;PDBTitle: crystal structure of the bacillus subtilis n-acetyltransferase ylbp2 protein in complex with coenzyme-a
4 c1y6zA_
13.6
16
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: heat shock protein, putative;PDBTitle: middle domain of plasmodium falciparum putative heat shock protein2 pf14_0417
5 c2jugB_
11.9
21
PDB header: biosynthetic proteinChain: B: PDB Molecule: tubc protein;PDBTitle: multienzyme docking in hybrid megasynthetases
6 d2gu2a1
11.1
23
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: AstE/AspA-like7 d1ci4a_
10.4
15
Fold: SAM domain-likeSuperfamily: Barrier-to-autointegration factor, BAFFamily: Barrier-to-autointegration factor, BAF8 c2f2iA_
10.1
50
PDB header: antimicrobial proteinChain: A: PDB Molecule: kalata-b1;PDBTitle: solution structure of [p20d,v21k]-kalata b1
9 c2kn8A_
9.4
24
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: dna cleavage and packaging protein large subunit, ul89;PDBTitle: nmr structure of the c-terminal domain of pul89
10 d3elga1
9.1
29
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like11 c3mdjB_
8.9
19
PDB header: hydrolase/hydrolase inhibitorChain: B: PDB Molecule: endoplasmic reticulum aminopeptidase 1;PDBTitle: er aminopeptidase, erap1, bound to the zinc aminopeptidase inhibitor,2 bestatin
12 c1kdxB_
8.4
40
PDB header: transcription regulation complexChain: B: PDB Molecule: creb;PDBTitle: kix domain of mouse cbp (creb binding protein) in complex2 with phosphorylated kinase inducible domain (pkid) of rat3 creb (cyclic amp response element binding protein), nmr 174 structures
13 d1cuka1
8.2
19
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain14 c3e4hA_
7.8
42
PDB header: plant proteinChain: A: PDB Molecule: varv peptide f;PDBTitle: crystal structure of the cyclotide varv f
15 c2jwmA_
7.6
33
PDB header: plant proteinChain: A: PDB Molecule: kalata-b7;PDBTitle: nmr spatial srtucture of ternary complex kalata b7/mn2+/dpc2 micelle
16 c3di3A_
7.5
19
PDB header: cytokine/cytokine receptorChain: A: PDB Molecule: interleukin-7;PDBTitle: crystal structure of the complex of human interleukin-7 with2 glycosylated human interleukin-7 receptor alpha ectodomain
17 d3duea1
6.8
25
Fold: BLIP-likeSuperfamily: BT0923-likeFamily: BT0923-like18 c2qfaC_
6.6
55
PDB header: cell cycle/cell cycle/cell cycleChain: C: PDB Molecule: inner centromere protein;PDBTitle: crystal structure of a survivin-borealin-incenp core complex
19 c2kzvA_
6.5
20
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution nmr structure of cv_0373(175-257) protein from2 chromobacterium violaceum, northeast structural genomics consortium3 target cvr118a
20 c3rjoA_
6.3
17
PDB header: hydrolaseChain: A: PDB Molecule: endoplasmic reticulum aminopeptidase 1;PDBTitle: crystal structure of erap1 peptide binding domain
21 d1pt4a_
not modelled
6.3
42
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Kalata B122 d1tyka_
not modelled
5.7
44
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: omega toxin-likeFamily: Spider toxins23 d1nb1a_
not modelled
5.5
50
Fold: Knottins (small inhibitors, toxins, lectins)Superfamily: CyclotidesFamily: Kalata B124 d2e1ba1
not modelled
5.4
24
Fold: Reductase/isomerase/elongation factor common domainSuperfamily: Translation proteinsFamily: AlaX-M N-terminal domain-like25 c1k0rB_
not modelled
5.3
13
PDB header: transcriptionChain: B: PDB Molecule: nusa;PDBTitle: crystal structure of mycobacterium tuberculosis nusa