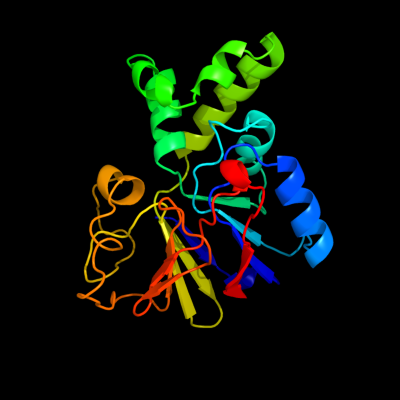
| 1 | d1g5ba_
|
|
 |
100.0 |
48 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
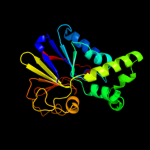
| 2 | c2qjcA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:diadenosine tetraphosphatase, putative;
PDBTitle: crystal structure of a putative diadenosine tetraphosphatase
|
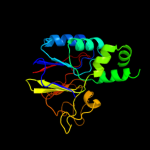
| 3 | c2dfjA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:diadenosinetetraphosphatase;
PDBTitle: crystal structure of the diadenosine tetraphosphate2 hydrolase from shigella flexneri 2a
|
| 4 | c2zbmA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein-tyrosine-phosphatase;
PDBTitle: crystal structure of i115m mutant cold-active protein2 tyrosine phosphatase
|
| 5 | d1jk7a_
|
|
 |
100.0 |
24 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 6 | d3c5wc1
|
|
 |
100.0 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 7 | d1s95a_
|
|
 |
100.0 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 8 | d1s70a_
|
|
 |
100.0 |
24 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 9 | c2jogA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:calmodulin-dependent calcineurin a subunit alpha
PDBTitle: structure of the calcineurin-nfat complex
|
| 10 | c3icfB_
|
|
 |
100.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:serine/threonine-protein phosphatase t;
PDBTitle: structure of protein serine/threonine phosphatase from saccharomyces2 cerevisiae with similarity to human phosphatase pp5
|
| 11 | d1auia_
|
|
 |
100.0 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 12 | c1auiA_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:serine/threonine phosphatase 2b;
PDBTitle: human calcineurin heterodimer
|
| 13 | d1nnwa_
|
|
 |
99.9 |
11 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Phosphoesterase-related |
| 14 | c1wao4_
|
|
 |
99.9 |
19 |
PDB header:hydrolase
Chain: 4: PDB Molecule:serine/threonine protein phosphatase 5;
PDBTitle: pp5 structure
|
| 15 | c2p6bC_
|
|
 |
99.9 |
19 |
PDB header:hydrolase/hydrolase regulator
Chain: C: PDB Molecule:calmodulin-dependent calcineurin a subunit alpha
PDBTitle: crystal structure of human calcineurin in complex with2 pvivit peptide
|
| 16 | d2p6ba1
|
|
 |
99.9 |
19 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Protein serine/threonine phosphatase |
| 17 | c3rqzC_
|
|
 |
99.9 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:metallophosphoesterase;
PDBTitle: crystal structure of metallophosphoesterase from sphaerobacter2 thermophilus
|
| 18 | d1uf3a_
|
|
 |
99.9 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
| 19 | c3qfnA_
|
|
 |
99.9 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
|
| 20 | c1su1A_
|
|
 |
99.9 |
25 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein yfce;
PDBTitle: structural and biochemical characterization of yfce, a2 phosphoesterase from e. coli
|
| 21 | d1su1a_ |
|
not modelled |
99.9 |
25 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 22 | d3ck2a1 |
|
not modelled |
99.9 |
20 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 23 | c2kknA_ |
|
not modelled |
99.8 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of themotoga maritima protein tm1076:2 northeast structural genomics consortium target vt57
|
| 24 | d2a22a1 |
|
not modelled |
99.8 |
19 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 25 | c1s3mA_ |
|
not modelled |
99.8 |
21 |
PDB header:phosphodiesterase
Chain: A: PDB Molecule:hypothetical protein mj0936;
PDBTitle: structural and functional characterization of a novel2 archaeal phosphodiesterase
|
| 26 | d1s3la_ |
|
not modelled |
99.8 |
21 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 27 | d2yvta1 |
|
not modelled |
99.8 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TT1561-like |
| 28 | d1z2wa1 |
|
not modelled |
99.7 |
22 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:YfcE-like |
| 29 | d1xm7a_ |
|
not modelled |
99.5 |
14 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Hypothetical protein aq 1666 |
| 30 | c3av0A_ |
|
not modelled |
99.5 |
15 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11-rad50 bound to atp s
|
| 31 | c3auzA_ |
|
not modelled |
99.4 |
15 |
PDB header:recombination
Chain: A: PDB Molecule:dna double-strand break repair protein mre11;
PDBTitle: crystal structure of mre11 with manganese
|
| 32 | d1ii7a_ |
|
not modelled |
99.4 |
19 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DNA double-strand break repair nuclease |
| 33 | c3rl4A_ |
|
not modelled |
99.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:metallophosphoesterase mpped2;
PDBTitle: rat metallophosphodiesterase mpped2 g252h mutant
|
| 34 | c3ib7A_ |
|
not modelled |
99.4 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:icc protein;
PDBTitle: crystal structure of full length rv0805
|
| 35 | d2hy1a1 |
|
not modelled |
99.3 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:GpdQ-like |
| 36 | c2hy1A_ |
|
not modelled |
99.3 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:rv0805;
PDBTitle: crystal structure of rv0805
|
| 37 | d3d03a1 |
|
not modelled |
99.2 |
10 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:GpdQ-like |
| 38 | c2xmoB_ |
|
not modelled |
99.1 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:lmo2642 protein;
PDBTitle: the crystal structure of lmo2642
|
| 39 | c2q8uA_ |
|
not modelled |
99.0 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:exonuclease, putative;
PDBTitle: crystal structure of mre11 from thermotoga maritima msb8 (tm1635) at2 2.20 a resolution
|
| 40 | d2nxfa1 |
|
not modelled |
99.0 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:ADPRibase-Mn-like |
| 41 | c1oidA_ |
|
not modelled |
98.8 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein usha;
PDBTitle: 5'-nucleotidase (e. coli) with an engineered disulfide2 bridge (s228c, p513c)
|
| 42 | c3qfkA_ |
|
not modelled |
98.8 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: 2.05 angstrom crystal structure of putative 5'-nucleotidase from2 staphylococcus aureus in complex with alpha-ketoglutarate
|
| 43 | d1xzwa2 |
|
not modelled |
98.8 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 44 | c3ivdA_ |
|
not modelled |
98.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleotidase;
PDBTitle: putative 5'-nucleotidase (c4898) from escherichia coli in2 complex with uridine
|
| 45 | d2qfra2 |
|
not modelled |
98.7 |
12 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 46 | d1usha2 |
|
not modelled |
98.6 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 47 | c3qg5D_ |
|
not modelled |
98.6 |
27 |
PDB header:hydrolase
Chain: D: PDB Molecule:mre11;
PDBTitle: the mre11:rad50 complex forms an atp dependent molecular clamp in dna2 double-strand break repair
|
| 48 | d1qhwa_ |
|
not modelled |
98.5 |
15 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 49 | c1qhwA_ |
|
not modelled |
98.5 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (purple acid phosphatase);
PDBTitle: purple acid phosphatase from rat bone
|
| 50 | c2z1aA_ |
|
not modelled |
98.5 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase precursor from thermus2 thermophilus hb8
|
| 51 | d1utea_ |
|
not modelled |
98.4 |
17 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:Purple acid phosphatase-like |
| 52 | d2z1aa2 |
|
not modelled |
98.4 |
16 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 53 | c3jyfB_ |
|
not modelled |
98.3 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:2',3'-cyclic nucleotide 2'-phosphodiesterase/3'-
PDBTitle: the crystal structure of a 2,3-cyclic nucleotide 2-2 phosphodiesterase/3-nucleotidase bifunctional periplasmic precursor3 protein from klebsiella pneumoniae subsp. pneumoniae mgh 78578
|
| 54 | c3t1iC_ |
|
not modelled |
98.3 |
30 |
PDB header:hydrolase
Chain: C: PDB Molecule:double-strand break repair protein mre11a;
PDBTitle: crystal structure of human mre11: understanding tumorigenic mutations
|
| 55 | c3gveB_ |
|
not modelled |
98.3 |
16 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:yfkn protein;
PDBTitle: crystal structure of calcineurin-like phosphoesterase yfkn from2 bacillus subtilis
|
| 56 | c3zu0A_ |
|
not modelled |
98.2 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:nad nucleotidase;
PDBTitle: structure of haemophilus influenzae nad nucleotidase (nadn)
|
| 57 | c2wdfA_ |
|
not modelled |
98.1 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:sulfur oxidation protein soxb;
PDBTitle: termus thermophilus sulfate thiohydrolase soxb
|
| 58 | d3c9fa2 |
|
not modelled |
97.0 |
23 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:5'-nucleotidase (syn. UDP-sugar hydrolase), N-terminal domain |
| 59 | c1xzwB_ |
|
not modelled |
96.7 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: sweet potato purple acid phosphatase/phosphate complex
|
| 60 | c1kbpB_ |
|
not modelled |
96.6 |
17 |
PDB header:hydrolase (phosphoric monoester)
Chain: B: PDB Molecule:purple acid phosphatase;
PDBTitle: kidney bean purple acid phosphatase
|
| 61 | c3c9fB_ |
|
not modelled |
96.2 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:5'-nucleotidase;
PDBTitle: crystal structure of 5'-nucleotidase from candida albicans sc5314
|
| 62 | d2z06a1 |
|
not modelled |
95.6 |
31 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:TTHA0625-like |
| 63 | c3e0jG_ |
|
not modelled |
95.2 |
15 |
PDB header:transferase
Chain: G: PDB Molecule:dna polymerase subunit delta-2;
PDBTitle: x-ray structure of the complex of regulatory subunits of2 human dna polymerase delta
|
| 64 | d1t71a_ |
|
not modelled |
95.1 |
22 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 65 | d1t70a_ |
|
not modelled |
95.0 |
21 |
Fold:Metallo-dependent phosphatases
Superfamily:Metallo-dependent phosphatases
Family:DR1281-like |
| 66 | c3floG_ |
|
not modelled |
91.7 |
20 |
PDB header:transferase
Chain: G: PDB Molecule:dna polymerase alpha subunit b;
PDBTitle: crystal structure of the carboxyl-terminal domain of yeast2 dna polymerase alpha in complex with its b subunit
|
| 67 | d1gg4a1 |
|
not modelled |
63.8 |
15 |
Fold:MurD-like peptide ligases, peptide-binding domain
Superfamily:MurD-like peptide ligases, peptide-binding domain
Family:MurCDEF C-terminal domain |
| 68 | c3nh8A_ |
|
not modelled |
47.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:aspartoacylase-2;
PDBTitle: crystal structure of murine aminoacylase 3 in complex with n-acetyl-s-2 1,2-dichlorovinyl-l-cysteine
|
| 69 | d1ylea1 |
|
not modelled |
35.0 |
29 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:AstA-like |
| 70 | c2issF_ |
|
not modelled |
29.4 |
21 |
PDB header:lyase, transferase
Chain: F: PDB Molecule:glutamine amidotransferase subunit pdxt;
PDBTitle: structure of the plp synthase holoenzyme from thermotoga maritima
|
| 71 | c2qj8B_ |
|
not modelled |
23.3 |
16 |
PDB header:hydrolase
Chain: B: PDB Molecule:mlr6093 protein;
PDBTitle: crystal structure of an aspartoacylase family protein (mlr6093) from2 mesorhizobium loti maff303099 at 2.00 a resolution
|
| 72 | c1z9bA_ |
|
not modelled |
22.6 |
19 |
PDB header:translation
Chain: A: PDB Molecule:translation initiation factor if-2;
PDBTitle: solution structure of the c1-subdomain of bacillus2 stearothermophilus translation initiation factor if2
|
| 73 | c3cdxB_ |
|
not modelled |
19.9 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:succinylglutamatedesuccinylase/aspartoacylase;
PDBTitle: crystal structure of2 succinylglutamatedesuccinylase/aspartoacylase from3 rhodobacter sphaeroides
|
| 74 | c3na6A_ |
|
not modelled |
19.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:succinylglutamate desuccinylase/aspartoacylase;
PDBTitle: crystal structure of a succinylglutamate desuccinylase (tm1040_2694)2 from silicibacter sp. tm1040 at 2.00 a resolution
|
| 75 | c2dlcX_ |
|
not modelled |
17.8 |
13 |
PDB header:ligase/trna
Chain: X: PDB Molecule:tyrosyl-trna synthetase, cytoplasmic;
PDBTitle: crystal structure of the ternary complex of yeast tyrosyl-trna2 synthetase
|
| 76 | d3beda1 |
|
not modelled |
15.2 |
11 |
Fold:PTS system fructose IIA component-like
Superfamily:PTS system fructose IIA component-like
Family:EIIA-man component-like |
| 77 | c3n9iA_ |
|
not modelled |
14.6 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from yersinia pestis2 co92
|
| 78 | c3prhB_ |
|
not modelled |
13.9 |
10 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: tryptophanyl-trna synthetase val144pro mutant from b. subtilis
|
| 79 | d1q7ra_ |
|
not modelled |
13.3 |
21 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 80 | c2el7A_ |
|
not modelled |
13.0 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from thermus2 thermophilus
|
| 81 | c2cybA_ |
|
not modelled |
12.9 |
17 |
PDB header:ligase
Chain: A: PDB Molecule:tyrosyl-trna synthetase;
PDBTitle: crystal structure of tyrosyl-trna synthetase complexed with2 l-tyrosine from archaeoglobus fulgidus
|
| 82 | d1j1ua_ |
|
not modelled |
12.2 |
27 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
| 83 | d1i6la_ |
|
not modelled |
12.1 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Class I aminoacyl-tRNA synthetases (RS), catalytic domain |
| 84 | d1otha2 |
|
not modelled |
12.0 |
12 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 85 | c3hp7A_ |
|
not modelled |
10.5 |
43 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hemolysin, putative;
PDBTitle: putative hemolysin from streptococcus thermophilus.
|
| 86 | d1ux6a1 |
|
not modelled |
10.1 |
33 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Thrombospondin C-terminal domain |
| 87 | c2vdwA_ |
|
not modelled |
10.0 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:vaccinia virus capping enzyme d1 subunit;
PDBTitle: guanosine n7 methyl-transferase sub-complex (d1-d12) of the2 vaccinia virus mrna capping enzyme
|
| 88 | d2at2a2 |
|
not modelled |
9.3 |
27 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 89 | d2z1ca1 |
|
not modelled |
8.6 |
40 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 90 | c3ujcA_ |
|
not modelled |
8.5 |
36 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoethanolamine n-methyltransferase;
PDBTitle: phosphoethanolamine methyltransferase mutant (h132a) from plasmodium2 falciparum in complex with phosphocholine
|
| 91 | d2o57a1 |
|
not modelled |
8.4 |
36 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
| 92 | d3d3ra1 |
|
not modelled |
7.8 |
30 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |
| 93 | c1gg4A_ |
|
not modelled |
7.8 |
14 |
PDB header:ligase
Chain: A: PDB Molecule:udp-n-acetylmuramoylalanyl-d-glutamyl-2,6-
PDBTitle: crystal structure of escherichia coli udpmurnac-tripeptide2 d-alanyl-d-alanine-adding enzyme (murf) at 2.3 angstrom3 resolution
|
| 94 | c3fmcC_ |
|
not modelled |
7.6 |
11 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative succinylglutamate desuccinylase / aspartoacylase;
PDBTitle: crystal structure of a putative succinylglutamate desuccinylase /2 aspartoacylase family protein (sama_0604) from shewanella amazonensis3 sb2b at 1.80 a resolution
|
| 95 | d1dxha2 |
|
not modelled |
7.5 |
12 |
Fold:ATC-like
Superfamily:Aspartate/ornithine carbamoyltransferase
Family:Aspartate/ornithine carbamoyltransferase |
| 96 | c3d3rA_ |
|
not modelled |
7.4 |
30 |
PDB header:chaperone
Chain: A: PDB Molecule:hydrogenase assembly chaperone hypc/hupf;
PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
|
| 97 | d2i3ca1 |
|
not modelled |
7.2 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:AstE/AspA-like |
| 98 | c3m5wB_ |
|
not modelled |
7.0 |
13 |
PDB header:ligase
Chain: B: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from2 campylobacter jejuni
|
| 99 | d2ot2a1 |
|
not modelled |
7.0 |
30 |
Fold:OB-fold
Superfamily:HupF/HypC-like
Family:HupF/HypC-like |