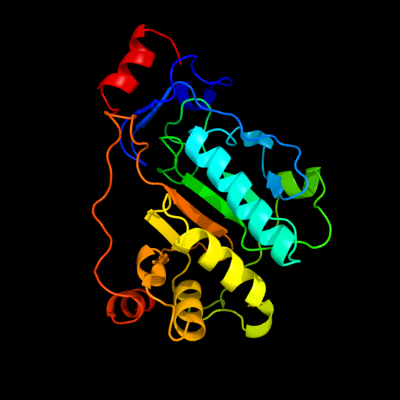
1 d1tzpa_
100.0
100
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: MepA-like2 c2vo9C_
80.3
17
PDB header: hydrolaseChain: C: PDB Molecule: l-alanyl-d-glutamate peptidase;PDBTitle: crystal structure of the enzymatically active domain of the2 listeria monocytogenes bacteriophage 500 endolysin ply500
3 d2vo9a1
41.6
16
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanY-like4 d1r44a_
39.2
17
Fold: Hedgehog/DD-peptidaseSuperfamily: Hedgehog/DD-peptidaseFamily: VanX-like5 c1lbuA_
33.4
36
PDB header: hydrolaseChain: A: PDB Molecule: muramoyl-pentapeptide carboxypeptidase;PDBTitle: hydrolase metallo (zn) dd-peptidase
6 c3ag5A_
25.2
22
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantothenate synthetase from staphylococcus2 aureus
7 d1ihoa_
23.3
39
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)8 d1v8fa_
22.4
30
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)9 c2faoB_
20.9
10
PDB header: hydrolase/transferaseChain: B: PDB Molecule: probable atp-dependent dna ligase;PDBTitle: crystal structure of pseudomonas aeruginosa ligd polymerase2 domain
10 c3innB_
20.7
35
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine-ligase in complex2 with atp at low occupancy at 2.1 a resolution
11 c3uk2B_
19.6
48
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
12 c2ejcA_
19.3
39
PDB header: ligaseChain: A: PDB Molecule: pantoate--beta-alanine ligase;PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
13 d2a84a1
18.6
35
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)14 c3n8hA_
18.4
48
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from francisella2 tularensis
15 c3mxtA_
15.5
44
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from campylobacter2 jejuni
16 c3c7bA_
15.0
33
PDB header: oxidoreductaseChain: A: PDB Molecule: sulfite reductase, dissimilatory-type subunit alpha;PDBTitle: structure of the dissimilatory sulfite reductase from archaeoglobus2 fulgidus
17 c2iruA_
14.8
8
PDB header: transferaseChain: A: PDB Molecule: putative dna ligase-like protein rv0938/mt0965;PDBTitle: crystal structure of the polymerase domain from mycobacterium2 tuberculosis ligase d
18 d2cwza1
14.8
4
Fold: Thioesterase/thiol ester dehydrase-isomeraseSuperfamily: Thioesterase/thiol ester dehydrase-isomeraseFamily: TTHA0967-like19 d1nkga3
12.7
22
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: Rhamnogalacturonase B, RhgB, N-terminal domain20 c1x9vA_
12.1
19
PDB header: viral proteinChain: A: PDB Molecule: vpr protein;PDBTitle: dimeric structure of the c-terminal domain of vpr
21 d1ujpa_
not modelled
12.0
19
Fold: TIM beta/alpha-barrelSuperfamily: Ribulose-phoshate binding barrelFamily: Tryptophan biosynthesis enzymes22 d2o8ra3
not modelled
11.8
32
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Polyphosphate kinase C-terminal domain23 c2jksA_
not modelled
11.4
17
PDB header: immune systemChain: A: PDB Molecule: bradyzoite surface antigen bsr4;PDBTitle: crystal structure of the the bradyzoite specific antigen2 bsr4 from toxoplasma gondii.
24 c2zuuA_
not modelled
9.8
18
PDB header: transferaseChain: A: PDB Molecule: lacto-n-biose phosphorylase;PDBTitle: crystal structure of galacto-n-biose/lacto-n-biose i phosphorylase in2 complex with glcnac
25 c3kuvB_
not modelled
9.7
8
PDB header: hydrolaseChain: B: PDB Molecule: fluoroacetyl coenzyme a thioesterase;PDBTitle: structural basis of the activity and substrate specificity of the2 fluoroacetyl-coa thioesterase flk - t42s mutant in complex with3 acetate.
26 d2v4ja2
not modelled
9.3
50
Fold: Ferredoxin-likeSuperfamily: Nitrite/Sulfite reductase N-terminal domain-likeFamily: DsrA/DsrB N-terminal-domain-like27 c3qooA_
not modelled
8.6
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of hot-dog-like taci_0573 protein from2 thermanaerovibrio acidaminovorans
28 d1gvia1
not modelled
7.3
19
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes29 c2x2zE_
not modelled
6.6
42
PDB header: membrane proteinChain: E: PDB Molecule: apical membrane antigen 1, putative;PDBTitle: crystal structure ama1 from toxoplasma gondii
30 c3navB_
not modelled
6.5
18
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase alpha chain;PDBTitle: crystal structure of an alpha subunit of tryptophan synthase from2 vibrio cholerae o1 biovar el tor str. n16961
31 d1wzla1
not modelled
6.4
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes32 d1pvta_
not modelled
6.0
17
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: AraD-like aldolase/epimerase33 d1j0ha1
not modelled
6.0
31
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: E-set domains of sugar-utilizing enzymes34 d1c25a_
not modelled
5.8
25
Fold: Rhodanese/Cell cycle control phosphataseSuperfamily: Rhodanese/Cell cycle control phosphataseFamily: Cell cycle control phosphatase, catalytic domain35 d1txla_
not modelled
5.6
29
Fold: LipocalinsSuperfamily: LipocalinsFamily: Hypothetical protein YodA36 c1txlA_
not modelled
5.6
29
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: metal-binding protein yoda;PDBTitle: crystal structure of metal-binding protein yoda from e.2 coli, pfam duf149
37 c2q8aA_
not modelled
5.5
33
PDB header: immune systemChain: A: PDB Molecule: apical membrane antigen 1;PDBTitle: structure of the malaria antigen ama1 in complex with a growth-2 inhibitory antibody
38 c2w82C_
not modelled
5.4
45
PDB header: replication inhibitorChain: C: PDB Molecule: orf18;PDBTitle: the structure of arda
39 d1dqaa1
not modelled
5.4
8
Fold: Ferredoxin-likeSuperfamily: NAD-binding domain of HMG-CoA reductaseFamily: NAD-binding domain of HMG-CoA reductase40 d1gqpa_
not modelled
5.3
27
Fold: Galactose-binding domain-likeSuperfamily: Galactose-binding domain-likeFamily: APC10-like41 c1h4uA_
not modelled
5.1
17
PDB header: extracellular matrix proteinChain: A: PDB Molecule: nidogen-1;PDBTitle: domain g2 of mouse nidogen-1
42 c2l57A_
not modelled
5.1
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: solution structure of an uncharacterized thioredoin-like protein from2 clostridium perfringens
43 d1xuba1
not modelled
5.0
17
Fold: Diaminopimelate epimerase-likeSuperfamily: Diaminopimelate epimerase-likeFamily: PhzC/PhzF-like