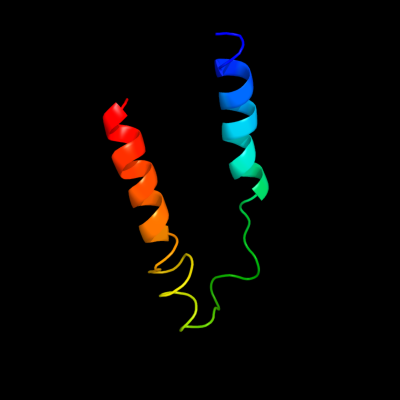
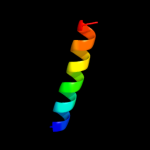
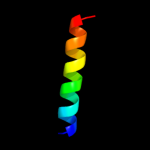
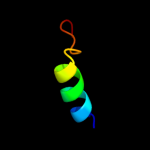
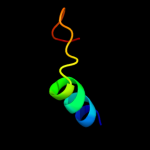
1 c3chxG_
59.5
30
PDB header: membrane proteinChain: G: PDB Molecule: pmoc;PDBTitle: crystal structure of methylosinus trichosporium ob3b2 particulate methane monooxygenase (pmmo)
2 d1prtf_
51.7
59
Fold: OB-foldSuperfamily: Bacterial enterotoxinsFamily: Bacterial AB5 toxins, B-subunits3 c1yewC_
44.7
30
PDB header: oxidoreductase, membrane proteinChain: C: PDB Molecule: particulate methane monooxygenase subunit c2;PDBTitle: crystal structure of particulate methane monooxygenase
4 d1pjwa_
25.8
67
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain5 d1s6na_
24.6
56
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain6 c3egpA_
24.6
33
PDB header: viral proteinChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure analysis of dengue-1 envelope protein2 domain iii
7 c3lbwB_
24.3
38
PDB header: transport proteinChain: B: PDB Molecule: m2 protein;PDBTitle: high resolution crystal structure of transmembrane domain of m2
8 c3lbwD_
24.3
38
PDB header: transport proteinChain: D: PDB Molecule: m2 protein;PDBTitle: high resolution crystal structure of transmembrane domain of m2
9 c3lbwC_
24.3
38
PDB header: transport proteinChain: C: PDB Molecule: m2 protein;PDBTitle: high resolution crystal structure of transmembrane domain of m2
10 c3lbwA_
24.3
38
PDB header: transport proteinChain: A: PDB Molecule: m2 protein;PDBTitle: high resolution crystal structure of transmembrane domain of m2
11 d1v32a_
19.4
23
Fold: SWIB/MDM2 domainSuperfamily: SWIB/MDM2 domainFamily: SWIB/MDM2 domain12 c2jqmA_
18.2
33
PDB header: transferaseChain: A: PDB Molecule: envelope protein e;PDBTitle: yellow fever envelope protein domain iii nmr structure2 (s288-k398)
13 c3c9jC_
18.2
38
PDB header: membrane proteinChain: C: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
14 c3c9jB_
18.2
38
PDB header: membrane proteinChain: B: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
15 c3c9jA_
18.2
38
PDB header: membrane proteinChain: A: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
16 c3c9jD_
18.2
38
PDB header: membrane proteinChain: D: PDB Molecule: proton channel protein m2, transmembrane segment;PDBTitle: the crystal structure of transmembrane domain of m2 protein and2 amantadine complex
17 d1v31a_
17.6
26
Fold: SWIB/MDM2 domainSuperfamily: SWIB/MDM2 domainFamily: SWIB/MDM2 domain18 d1ixra1
14.6
43
Fold: SAM domain-likeSuperfamily: RuvA domain 2-likeFamily: DNA helicase RuvA subunit, middle domain19 c2h0pA_
14.0
22
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: nmr structure of the dengue-4 virus envelope protein domain2 iii
20 d1uhra_
12.6
27
Fold: SWIB/MDM2 domainSuperfamily: SWIB/MDM2 domainFamily: SWIB/MDM2 domain21 c2kncA_
not modelled
12.1
31
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
22 d2iuba2
not modelled
11.9
17
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region23 c2wwbC_
not modelled
11.9
36
PDB header: ribosomeChain: C: PDB Molecule: protein transport protein sec61 subunit beta;PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
24 c1nyjB_
not modelled
9.6
33
PDB header: viral proteinChain: B: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
25 c2kqtC_
not modelled
9.6
33
PDB header: transport proteinChain: C: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
26 c2kqtA_
not modelled
9.6
33
PDB header: transport proteinChain: A: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
27 c1nyjA_
not modelled
9.6
33
PDB header: viral proteinChain: A: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
28 c1nyjD_
not modelled
9.6
33
PDB header: viral proteinChain: D: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
29 c2kqtB_
not modelled
9.6
33
PDB header: transport proteinChain: B: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
30 c1nyjC_
not modelled
9.6
33
PDB header: viral proteinChain: C: PDB Molecule: matrix protein m2;PDBTitle: the closed state structure of m2 protein h+ channel by2 solid state nmr spectroscopy
31 c1mp6A_
not modelled
9.6
33
PDB header: membrane proteinChain: A: PDB Molecule: matrix protein m2;PDBTitle: structure of the transmembrane region of the m2 protein h+2 channel by solid state nmr spectroscopy
32 c2kqtD_
not modelled
9.6
33
PDB header: transport proteinChain: D: PDB Molecule: m2 protein;PDBTitle: solid-state nmr structure of the m2 transmembrane peptide of the2 influenza a virus in dmpc lipid bilayers bound to deuterated3 amantadine
33 c2bbjB_
not modelled
9.0
14
PDB header: metal transport/membrane proteinChain: B: PDB Molecule: divalent cation transport-related protein;PDBTitle: crystal structure of the cora mg2+ transporter
34 d2fp7b1
not modelled
8.3
55
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases35 c3bkdE_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: E: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
36 c3bkdG_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: G: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
37 c3bkdH_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: H: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
38 c3bkdD_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: D: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
39 c3bkdC_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: C: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
40 c3bkdA_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: A: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
41 c3bkdF_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: F: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
42 c3bkdB_
not modelled
8.1
33
PDB header: viral protein, membrane proteinChain: B: PDB Molecule: transmembrane domain of matrix protein m2;PDBTitle: high resolution crystal structure of transmembrane domain of m22 protein
43 c3c6dB_
not modelled
7.1
33
PDB header: virusChain: B: PDB Molecule: polyprotein;PDBTitle: the pseudo-atomic structure of dengue immature virus
44 c2ehoL_
not modelled
6.8
16
PDB header: replicationChain: L: PDB Molecule: gins complex subunit 3;PDBTitle: crystal structure of human gins complex
45 c3i9yA_
not modelled
6.7
13
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of the v. parahaemolyticus histidine2 kinase sensor tors sensor domain
46 d1ok8a1
not modelled
6.5
38
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain47 d2fomb1
not modelled
6.2
55
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases48 c3e90B_
not modelled
6.2
55
PDB header: hydrolaseChain: B: PDB Molecule: ns3 protease;PDBTitle: west nile vi rus ns2b-ns3protease in complexed with2 inhibitor naph-kkr-h
49 c2rlfA_
not modelled
6.2
33
PDB header: proton transportChain: A: PDB Molecule: matrix protein 2;PDBTitle: proton channel m2 from influenza a in complex with2 inhibitor rimantadine
50 d2ijob1
not modelled
6.2
55
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases51 c3uajA_
not modelled
5.8
22
PDB header: viral protein/immune systemChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure of the envelope glycoprotein ectodomain from dengue2 virus serotype 4 in complex with the fab fragment of the chimpanzee3 monoclonal antibody 5h2
52 c2w8xB_
not modelled
5.7
40
PDB header: membrane proteinChain: B: PDB Molecule: ion-channel modulator raklp;PDBTitle: structure of the tick ion-channel modulator ra-klp
53 c1urzC_
not modelled
5.6
44
PDB header: virus/viral proteinChain: C: PDB Molecule: envelope protein;PDBTitle: low ph induced, membrane fusion conformation of the2 envelope protein of tick-borne encephalitis virus
54 d2f2ab1
not modelled
5.5
39
Fold: GatB/YqeY motifSuperfamily: GatB/YqeY motifFamily: GatB/GatE C-terminal domain-like55 d1befa_
not modelled
5.4
55
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases56 c3lkwA_
not modelled
5.4
55
PDB header: viral protein,hydrolaseChain: A: PDB Molecule: fusion protein of nonstructural protein 2b andPDBTitle: crystal structure of dengue virus 1 ns2b/ns3 protease active2 site mutant
57 d1ztxe1
not modelled
5.3
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Class II viral fusion proteins C-terminal domain58 c2kadD_
not modelled
5.3
29
PDB header: membrane proteinChain: D: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
59 c2kadA_
not modelled
5.3
29
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
60 c2kadC_
not modelled
5.3
29
PDB header: membrane proteinChain: C: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain
61 c2kadB_
not modelled
5.3
29
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane peptide of matrix protein 2;PDBTitle: magic-angle-spinning solid-state nmr structure of influenza2 a m2 transmembrane domain