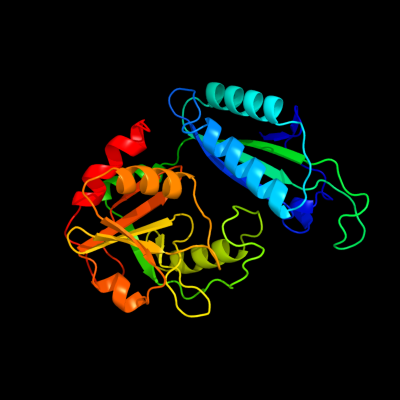
| 1 | d1i2ka_
|
|
 |
100.0 |
100 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
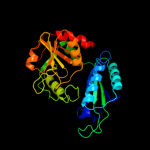
| 2 | c2xpfB_
|
|
 |
100.0 |
33 |
PDB header:lyase
Chain: B: PDB Molecule:4-amino-4-deoxychorismate lyase;
PDBTitle: crystal structure of putative aminodeoxychorismate lyase2 from pseudomonas aeruginosa
|
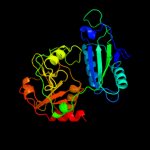
| 3 | c1wrvB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:branched-chain amino acid aminotransferase;
PDBTitle: crystal structure of t.th.hb8 branched-chain amino acid2 aminotransferase
|
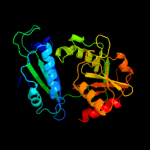
| 4 | d1iyea_
|
|
 |
100.0 |
21 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
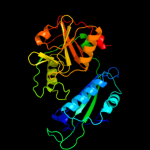
| 5 | d1daaa_
|
|
 |
100.0 |
21 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
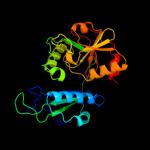
| 6 | c3lulA_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:4-amino-4-deoxychorismate lyase;
PDBTitle: crystal structure of putative 4-amino-4-deoxychorismate lyase.2 (yp_094631.1) from legionella pneumophila subsp. pneumophila str.3 philadelphia 1 at 1.78 a resolution
|
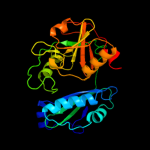
| 7 | c3cswB_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: B: PDB Molecule:putative branched-chain-amino-acid aminotransferase;
PDBTitle: crystal structure of a putative branched-chain amino acid2 aminotransferase (tm0831) from thermotoga maritima at 2.15 a3 resolution
|
| 8 | c3dtfB_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:branched-chain amino acid aminotransferase;
PDBTitle: structural analysis of mycobacterial branched chain aminotransferase-2 implications for inhibitor design
|
| 9 | d2a1ha1
|
|
 |
100.0 |
15 |
Fold:D-aminoacid aminotransferase-like PLP-dependent enzymes
Superfamily:D-aminoacid aminotransferase-like PLP-dependent enzymes
Family:D-aminoacid aminotransferase-like PLP-dependent enzymes |
| 10 | c2zgiA_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:putative 4-amino-4-deoxychorismate lyase;
PDBTitle: crystal structure of putative 4-amino-4-deoxychorismate lyase
|
| 11 | c2abjG_
|
|
 |
100.0 |
15 |
PDB header:transferase
Chain: G: PDB Molecule:branched-chain-amino-acid aminotransferase, cytosolic;
PDBTitle: crystal structure of human branched chain amino acid transaminase in a2 complex with an inhibitor, c16h10n2o4f3scl, and pyridoxal 5'3 phosphate.
|
| 12 | c3snoA_
|
|
 |
100.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:hypothetical aminotransferase;
PDBTitle: crystal structure of a hypothetical aminotransferase (ncgl2491) from2 corynebacterium glutamicum atcc 13032 kitasato at 1.60 a resolution
|
| 13 | c3qqmD_
|
|
 |
100.0 |
22 |
PDB header:transferase
Chain: D: PDB Molecule:mlr3007 protein;
PDBTitle: crystal structure of a putative amino-acid aminotransferase2 (np_104211.1) from mesorhizobium loti at 2.30 a resolution
|
| 14 | c3cebA_
|
|
 |
100.0 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:d-aminoacid aminotransferase-like plp-dependent enzyme;
PDBTitle: crystal structure of a putative 4-amino-4-deoxychorismate lyase2 (hs_0128) from haemophilus somnus 129pt at 2.40 a resolution
|
| 15 | d1ljra2
|
|
 |
64.7 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 16 | d1jlva2
|
|
 |
54.6 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 17 | d1oyja2
|
|
 |
53.5 |
20 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 18 | d1aw9a2
|
|
 |
52.4 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 19 | d1axda2
|
|
 |
44.1 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 20 | d1r5aa2
|
|
 |
41.9 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 21 | d1pn9a2 |
|
not modelled |
38.2 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 22 | c2klxA_ |
|
not modelled |
36.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glutaredoxin;
PDBTitle: solution structure of glutaredoxin from bartonella henselae str.2 houston
|
| 23 | d1gnwa2 |
|
not modelled |
33.8 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 24 | d1e6ba2 |
|
not modelled |
32.8 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 25 | d1k3ya2 |
|
not modelled |
32.5 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 26 | d1v2aa2 |
|
not modelled |
27.1 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 27 | c2cb1A_ |
|
not modelled |
27.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:o-acetyl homoserine sulfhydrylase;
PDBTitle: crystal structure of o-actetyl homoserine sulfhydrylase2 from thermus thermophilus hb8,oah2.
|
| 28 | d1rxxa_ |
|
not modelled |
22.0 |
18 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Arginine deiminase |
| 29 | d2fnoa2 |
|
not modelled |
22.0 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 30 | d1tw9a2 |
|
not modelled |
20.8 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 31 | d1fhea2 |
|
not modelled |
18.5 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 32 | d1ev4a2 |
|
not modelled |
18.1 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 33 | c8jdwA_ |
|
not modelled |
17.9 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:protein (l-arginine:glycine amidinotransferase);
PDBTitle: crystal structure of human l-arginine:glycine2 amidinotransferase in complex with l-alanine
|
| 34 | d1jdwa_ |
|
not modelled |
16.9 |
5 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Amidinotransferase |
| 35 | c1jdwA_ |
|
not modelled |
16.9 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:l-arginine\:glycine amidinotransferase;
PDBTitle: crystal structure and mechanism of l-arginine: glycine2 amidinotransferase: a mitochondrial enzyme involved in3 creatine biosynthesis
|
| 36 | d1tu7a2 |
|
not modelled |
16.9 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 37 | d1gwca2 |
|
not modelled |
16.4 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 38 | c3kgkA_ |
|
not modelled |
15.8 |
12 |
PDB header:chaperone
Chain: A: PDB Molecule:arsenical resistance operon trans-acting repressor arsd;
PDBTitle: crystal structure of arsd
|
| 39 | d2gk3a1 |
|
not modelled |
15.8 |
21 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:STM3548-like |
| 40 | d1pd212 |
|
not modelled |
15.4 |
11 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 41 | c3ktbD_ |
|
not modelled |
15.4 |
7 |
PDB header:transcription regulator
Chain: D: PDB Molecule:arsenical resistance operon trans-acting repressor;
PDBTitle: crystal structure of arsenical resistance operon trans-acting2 repressor from bacteroides vulgatus atcc 8482
|
| 42 | d1r7ha_ |
|
not modelled |
15.0 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 43 | d1nm3a1 |
|
not modelled |
14.6 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 44 | d2gsta2 |
|
not modelled |
14.5 |
19 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 45 | d1jlwa2 |
|
not modelled |
14.5 |
13 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 46 | d1h75a_ |
|
not modelled |
14.0 |
18 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Thioltransferase |
| 47 | d1xw6a2 |
|
not modelled |
13.7 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 48 | c3h1nA_ |
|
not modelled |
13.3 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:probable glutathione s-transferase;
PDBTitle: crystal structure of probable glutathione s-transferase from2 bordetella bronchiseptica rb50
|
| 49 | d1t1va_ |
|
not modelled |
13.2 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:SH3BGR (SH3-binding, glutamic acid-rich protein-like) |
| 50 | c3ndnC_ |
|
not modelled |
13.1 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:o-succinylhomoserine sulfhydrylase;
PDBTitle: crystal structure of o-succinylhomoserine sulfhydrylase from2 mycobacterium tuberculosis covalently bound to pyridoxal-5-phosphate
|
| 51 | d1fw1a2 |
|
not modelled |
12.8 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 52 | d1glqa2 |
|
not modelled |
12.6 |
18 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 53 | d2fhea2 |
|
not modelled |
12.2 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 54 | d1duga2 |
|
not modelled |
11.4 |
21 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 55 | d1bwda_ |
|
not modelled |
11.0 |
21 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Amidinotransferase |
| 56 | c2hueB_ |
|
not modelled |
10.8 |
9 |
PDB header:dna binding protein
Chain: B: PDB Molecule:histone h3;
PDBTitle: structure of the h3-h4 chaperone asf1 bound to histones h3 and h4
|
| 57 | d2c4ja2 |
|
not modelled |
10.7 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 58 | d2nv0a1 |
|
not modelled |
10.5 |
13 |
Fold:Flavodoxin-like
Superfamily:Class I glutamine amidotransferase-like
Family:Class I glutamine amidotransferases (GAT) |
| 59 | c2nmpC_ |
|
not modelled |
10.4 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine gamma-lyase;
PDBTitle: crystal structure of human cystathionine gamma lyase
|
| 60 | d1rk4a2 |
|
not modelled |
10.1 |
14 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 61 | d1s9ra_ |
|
not modelled |
9.9 |
11 |
Fold:Pentein, beta/alpha-propeller
Superfamily:Pentein
Family:Arginine deiminase |
| 62 | d2ftsa3 |
|
not modelled |
9.8 |
11 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MoeA central domain-like |
| 63 | d2a2ra2 |
|
not modelled |
9.7 |
20 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 64 | d2gsqa2 |
|
not modelled |
9.5 |
17 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 65 | c1ua5A_ |
|
not modelled |
9.4 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:glutathione s-transferase;
PDBTitle: non-fusion gst from s. japonicum in complex with glutathione
|
| 66 | d2f99a1 |
|
not modelled |
9.0 |
11 |
Fold:Cystatin-like
Superfamily:NTF2-like
Family:SnoaL-like polyketide cyclase |
| 67 | d1kx5a_ |
|
not modelled |
9.0 |
7 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 68 | d1k0da2 |
|
not modelled |
9.0 |
8 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 69 | d1p3ie_ |
|
not modelled |
8.8 |
9 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 70 | c1ibjC_ |
|
not modelled |
8.7 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:cystathionine beta-lyase;
PDBTitle: crystal structure of cystathionine beta-lyase from arabidopsis2 thaliana
|
| 71 | c2c5sA_ |
|
not modelled |
8.7 |
15 |
PDB header:rna-binding protein
Chain: A: PDB Molecule:probable thiamine biosynthesis protein thii;
PDBTitle: crystal structure of bacillus anthracis thii, a trna-2 modifying enzyme containing the predicted rna-binding3 thump domain
|
| 72 | c2jl4A_ |
|
not modelled |
8.6 |
7 |
PDB header:isomerase
Chain: A: PDB Molecule:maleylpyruvate isomerase;
PDBTitle: holo structure of maleyl pyruvate isomerase, a bacterial2 glutathione-s-transferase in zeta class
|
| 73 | d2c5sa1 |
|
not modelled |
8.5 |
15 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ThiI-like |
| 74 | d2cvda2 |
|
not modelled |
8.0 |
11 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 75 | d2gsra2 |
|
not modelled |
7.9 |
19 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 76 | d1dzfa1 |
|
not modelled |
7.9 |
5 |
Fold:Restriction endonuclease-like
Superfamily:Eukaryotic RPB5 N-terminal domain
Family:Eukaryotic RPB5 N-terminal domain |
| 77 | d1b48a2 |
|
not modelled |
7.8 |
16 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 78 | c1y1yE_ |
|
not modelled |
7.8 |
6 |
PDB header:transferase/transcription/dna-rna hybrid
Chain: E: PDB Molecule:dna-directed rna polymerases i, ii, and iii 27
PDBTitle: rna polymerase ii-tfiis-dna/rna complex
|
| 79 | d1m0ua2 |
|
not modelled |
7.7 |
19 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 80 | c3b4oB_ |
|
not modelled |
7.7 |
14 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:phenazine biosynthesis protein a/b;
PDBTitle: crystal structure of phenazine biosynthesis protein phza/b2 from burkholderia cepacia r18194, apo form
|
| 81 | c1htmB_ |
|
not modelled |
7.4 |
50 |
PDB header:viral protein
Chain: B: PDB Molecule:hemagglutinin ha2 chain;
PDBTitle: structure of influenza haemagglutinin at the ph of membrane2 fusion
|
| 82 | d1eqzg_ |
|
not modelled |
7.4 |
9 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 83 | d1k0dd2 |
|
not modelled |
7.2 |
6 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 84 | d1ng7a_ |
|
not modelled |
6.8 |
8 |
Fold:Soluble domain of poliovirus core protein 3a
Superfamily:Soluble domain of poliovirus core protein 3a
Family:Soluble domain of poliovirus core protein 3a |
| 85 | c2kppA_ |
|
not modelled |
6.7 |
8 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:lin0431 protein;
PDBTitle: solution nmr structure of lin0431 protein from listeria innocua.2 northeast structural genomics consortium target lkr112
|
| 86 | c3ff0A_ |
|
not modelled |
6.6 |
20 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:phenazine biosynthesis protein phzb 2;
PDBTitle: crystal structure of a phenazine biosynthesis-related protein (phzb2)2 from pseudomonas aeruginosa at 1.90 a resolution
|
| 87 | c1tw9C_ |
|
not modelled |
6.6 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:glutathione s-transferase 2;
PDBTitle: glutathione transferase-2, apo form, from the nematode heligmosomoides2 polygyrus
|
| 88 | d1ibja_ |
|
not modelled |
6.5 |
12 |
Fold:PLP-dependent transferase-like
Superfamily:PLP-dependent transferases
Family:Cystathionine synthase-like |
| 89 | c3lkxA_ |
|
not modelled |
6.5 |
22 |
PDB header:chaperone
Chain: A: PDB Molecule:transcription factor btf3;
PDBTitle: human nac dimerization domain
|
| 90 | d1ygya4 |
|
not modelled |
6.4 |
18 |
Fold:FwdE/GAPDH domain-like
Superfamily:Serine metabolism enzymes domain
Family:SerA intervening domain-like |
| 91 | c2k6tA_ |
|
not modelled |
6.2 |
21 |
PDB header:hormone
Chain: A: PDB Molecule:insulin-like 3 a chain;
PDBTitle: solution structure of the relaxin-like factor
|
| 92 | d1id3a_ |
|
not modelled |
6.2 |
6 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 93 | d1gula2 |
|
not modelled |
6.2 |
15 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
| 94 | d2hqya2 |
|
not modelled |
6.1 |
7 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:FemXAB nonribosomal peptidyltransferases |
| 95 | d1tzyc_ |
|
not modelled |
6.0 |
7 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
| 96 | c1gumA_ |
|
not modelled |
6.0 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:protein (glutathione transferase a4-4);
PDBTitle: human glutathione transferase a4-4 without ligands
|
| 97 | d1vk9a_ |
|
not modelled |
5.9 |
11 |
Fold:Cytidine deaminase-like
Superfamily:Cytidine deaminase-like
Family:Hypothetical protein TM1506 |
| 98 | d1vbka1 |
|
not modelled |
5.8 |
6 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Adenine nucleotide alpha hydrolases-like
Family:ThiI-like |
| 99 | d2a1ja1 |
|
not modelled |
5.8 |
16 |
Fold:SAM domain-like
Superfamily:RuvA domain 2-like
Family:Hef domain-like |