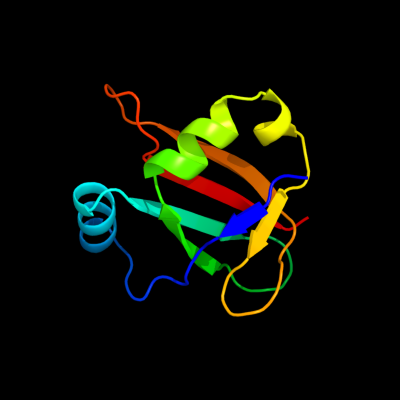
1 d1dfup_
100.0
100
Fold: Ribosomal protein L25-likeSuperfamily: Ribosomal protein L25-likeFamily: Ribosomal protein L25-like2 d1feua_
100.0
18
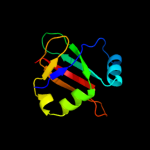
Fold: Ribosomal protein L25-likeSuperfamily: Ribosomal protein L25-likeFamily: Ribosomal protein L25-like3 d2zjrs1
99.9
19
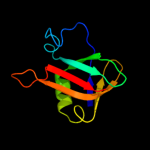
Fold: Ribosomal protein L25-likeSuperfamily: Ribosomal protein L25-likeFamily: Ribosomal protein L25-like4 c2b9nZ_
99.9
20
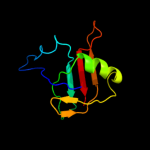
PDB header: ribosomeChain: Z: PDB Molecule: 50s ribosomal protein ctc;PDBTitle: 50s ribosomal subunit from a crystal structure of release factor rf2,2 trnas and mrna bound to the ribosome. this file contains the 50s3 subunit from a crystal structure of release factor rf1, trnas and4 mrna bound to the ribosome and is described in remark 400.
5 d1zaka2
52.9
25
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain6 d1htwa_
47.9
16
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: YjeE-like7 c1vhoA_
24.0
8
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: endoglucanase;PDBTitle: crystal structure of a putative peptidase/endoglucanase
8 c1q7lB_
18.0
25
PDB header: hydrolaseChain: B: PDB Molecule: aminoacylase-1;PDBTitle: zn-binding domain of the t347g mutant of human aminoacylase-2 i
9 c3no8A_
14.8
20
PDB header: isomerase regulatorChain: A: PDB Molecule: btb/poz domain-containing protein 2;PDBTitle: crystal structure of the phr domain from human btbd2 protein
10 c1vixA_
14.8
14
PDB header: hydrolaseChain: A: PDB Molecule: peptidase t;PDBTitle: crystal structure of a putative peptidase t
11 d2phpa1
12.8
29
Fold: AraD/HMP-PK domain-likeSuperfamily: AraD/HMP-PK domain-likeFamily: Phosphomethylpyrimidine kinase C-terminal domain-like12 d1a6ca3
12.2
10
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Positive stranded ssRNA virusesFamily: Comoviridae-like VP13 c3hwjA_
11.2
33
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase mycbp2;PDBTitle: crystal structure of the second phr domain of mouse myc-2 binding protein 2 (mycbp-2)
14 c3l7pA_
11.0
17
PDB header: transcriptionChain: A: PDB Molecule: putative nitrogen regulatory protein pii;PDBTitle: crystal structure of smu.1657c, putative nitrogen regulatory protein2 pii from streptococcus mutans
15 d1etha1
10.6
14
Fold: Lipase/lipooxygenase domain (PLAT/LH2 domain)Superfamily: Lipase/lipooxygenase domain (PLAT/LH2 domain)Family: Colipase-binding domain16 c3rzaA_
9.6
10
PDB header: hydrolaseChain: A: PDB Molecule: tripeptidase;PDBTitle: crystal structure of a tripeptidase (sav1512) from staphylococcus2 aureus subsp. aureus mu50 at 2.10 a resolution
17 c3na2C_
9.5
18
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of protein of unknown function from mine drainage2 metagenome leptospirillum rubarum
18 c3ls8A_
9.3
67
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol 3-kinase catalytic subunitPDBTitle: crystal structure of human pik3c3 in complex with 3-[4-(4-2 morpholinyl)thieno[3,2-d]pyrimidin-2-yl]-phenol
19 d1z2la1
9.3
10
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases20 d1vjna_
8.9
24
Fold: Metallo-hydrolase/oxidoreductaseSuperfamily: Metallo-hydrolase/oxidoreductaseFamily: Hypothetical protein TM020721 c3gbwA_
not modelled
8.8
33
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase mycbp2;PDBTitle: crystal structure of the first phr domain of the mouse myc-2 binding protein 2 (mycbp-2)
22 c2j8aA_
not modelled
8.6
14
PDB header: transferaseChain: A: PDB Molecule: histone-lysine n-methyltransferase, h3 lysine-4PDBTitle: x-ray structure of the n-terminus rrm domain of set1
23 d1e7ua4
not modelled
8.3
56
Fold: Protein kinase-like (PK-like)Superfamily: Protein kinase-like (PK-like)Family: Phoshoinositide 3-kinase (PI3K), catalytic domain24 c2x6kB_
not modelled
7.5
67
PDB header: transferaseChain: B: PDB Molecule: phosphotidylinositol 3 kinase 59f;PDBTitle: the crystal structure of the drosophila class iii pi3-kinase2 vps34 in complex with pi-103
25 c3ifeA_
not modelled
7.4
11
PDB header: hydrolaseChain: A: PDB Molecule: peptidase t;PDBTitle: 1.55 angstrom resolution crystal structure of peptidase t (pept-1)2 from bacillus anthracis str. 'ames ancestor'.
26 d1qo5b_
not modelled
7.1
29
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase27 c3mmtC_
not modelled
7.0
26
PDB header: hydrolaseChain: C: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: crystal structure of fructose bisphosphate aldolase from bartonella2 henselae, bound to fructose bisphosphate
28 d1fdja_
not modelled
6.6
29
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase29 d1vixa1
not modelled
6.4
14
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: Bacterial dinuclear zinc exopeptidases30 d1xfba1
not modelled
6.4
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase31 d1vkia_
not modelled
6.3
5
Fold: YbaK/ProRS associated domainSuperfamily: YbaK/ProRS associated domainFamily: YbaK/ProRS associated domain32 d1zaia1
not modelled
6.2
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase33 c2x0oA_
not modelled
6.2
21
PDB header: biosynthetic proteinChain: A: PDB Molecule: alcaligin biosynthesis protein;PDBTitle: apo structure of the alcaligin biosynthesis protein c (alcc)2 from bordetella bronchiseptica
34 c2pc4B_
not modelled
6.0
19
PDB header: lyaseChain: B: PDB Molecule: fructose-bisphosphate aldolase;PDBTitle: crystal structure of fructose-bisphosphate aldolase from plasmodium2 falciparum in complex with trap-tail determined at 2.4 angstrom3 resolution
35 d1a5ca_
not modelled
5.7
19
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase36 c2rb7A_
not modelled
5.7
19
PDB header: hydrolaseChain: A: PDB Molecule: peptidase, m20/m25/m40 family;PDBTitle: crystal structure of co-catalytic metallopeptidase (yp_387682.1) from2 desulfovibrio desulfuricans g20 at 1.60 a resolution
37 d1fbaa_
not modelled
5.7
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase38 d1f2ja_
not modelled
5.4
23
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase39 d1nn7a_
not modelled
5.3
17
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels40 c2y3aA_
not modelled
5.1
56
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol-4,5-bisphosphate 3-kinase catalyticPDBTitle: crystal structure of p110beta in complex with icsh2 of p85beta and2 the drug gdc-0941
41 c3ic1A_
not modelled
5.1
9
PDB header: hydrolaseChain: A: PDB Molecule: succinyl-diaminopimelate desuccinylase;PDBTitle: crystal structure of zinc-bound succinyl-diaminopimelate desuccinylase2 from haemophilus influenzae
42 d1s1ga_
not modelled
5.1
19
Fold: POZ domainSuperfamily: POZ domainFamily: Tetramerization domain of potassium channels43 c1e8zA_
not modelled
5.0
56
PDB header: transferaseChain: A: PDB Molecule: phosphatidylinositol 3-kinase catalytic subunit;PDBTitle: structure determinants of phosphoinositide 3-kinase2 inhibition by wortmannin, ly294002, quercetin, myricetin3 and staurosporine
44 d2hgca1
not modelled
5.0
16
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: YjcQ-like