| 1 |
|
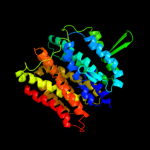
PDB 1pw4 chain A
Region: 1 - 412
Aligned: 392
Modelled: 406
Confidence: 100.0%
Identity: 17%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: Glycerol-3-phosphate transporter
Phyre2
| 2 |
|
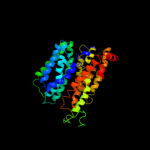
PDB 3o7p chain A
Region: 3 - 399
Aligned: 375
Modelled: 397
Confidence: 99.9%
Identity: 13%
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
Phyre2
| 3 |
|
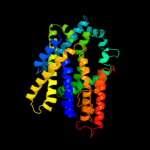
PDB 1pv7 chain A
Region: 1 - 411
Aligned: 396
Modelled: 411
Confidence: 99.9%
Identity: 11%
Fold: MFS general substrate transporter
Superfamily: MFS general substrate transporter
Family: LacY-like proton/sugar symporter
Phyre2
| 4 |
|
PDB 2gfp chain A
Region: 1 - 388
Aligned: 356
Modelled: 385
Confidence: 99.9%
Identity: 15%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 5 |
|
PDB 2xut chain C
Region: 19 - 395
Aligned: 362
Modelled: 377
Confidence: 99.9%
Identity: 12%
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
Phyre2
| 6 |
|
PDB 3qnq chain D
Region: 362 - 414
Aligned: 53
Modelled: 53
Confidence: 75.1%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
Phyre2
| 7 |
|
PDB 2xq2 chain A
Region: 48 - 415
Aligned: 363
Modelled: 368
Confidence: 26.8%
Identity: 10%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 8 |
|
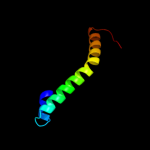
PDB 2g9p chain A
Region: 48 - 61
Aligned: 14
Modelled: 14
Confidence: 18.6%
Identity: 43%
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
Phyre2
| 9 |
|
PDB 3ff5 chain B
Region: 398 - 414
Aligned: 17
Modelled: 17
Confidence: 10.8%
Identity: 12%
PDB header:protein transport
Chain: B: PDB Molecule:peroxisomal biogenesis factor 14;
PDBTitle: crystal structure of the conserved n-terminal domain of the2 peroxisomal matrix-protein-import receptor, pex14p
Phyre2
| 10 |
|
PDB 2wwa chain A
Region: 373 - 425
Aligned: 53
Modelled: 53
Confidence: 10.4%
Identity: 11%
PDB header:ribosome
Chain: A: PDB Molecule:sec sixty-one protein homolog;
PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the2 yeast 80s ribosome
Phyre2
| 11 |
|
PDB 2wwb chain A
Region: 373 - 426
Aligned: 54
Modelled: 54
Confidence: 8.9%
Identity: 13%
PDB header:ribosome
Chain: A: PDB Molecule:protein transport protein sec61 subunit alpha isoform 1;
PDBTitle: cryo-em structure of the mammalian sec61 complex bound to the2 actively translating wheat germ 80s ribosome
Phyre2
| 12 |
|
PDB 2w85 chain A
Region: 398 - 419
Aligned: 22
Modelled: 22
Confidence: 7.7%
Identity: 9%
PDB header:protein transport
Chain: A: PDB Molecule:peroxisomal membrane anchor protein pex14;
PDBTitle: structure of pex14 in compex with pex19
Phyre2
| 13 |
|
PDB 2inp chain D
Region: 49 - 60
Aligned: 12
Modelled: 12
Confidence: 6.3%
Identity: 33%
PDB header:oxidoreductase
Chain: D: PDB Molecule:phenol hydroxylase component phl;
PDBTitle: structure of the phenol hydroxylase-regulatory protein2 complex
Phyre2
| 14 |
|
PDB 3hd6 chain A
Region: 198 - 422
Aligned: 220
Modelled: 225
Confidence: 6.2%
Identity: 9%
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
Phyre2
| 15 |
|
PDB 3mmy chain F
Region: 395 - 408
Aligned: 14
Modelled: 14
Confidence: 5.9%
Identity: 29%
PDB header:nuclear protein
Chain: F: PDB Molecule:nuclear pore complex protein nup98;
PDBTitle: structural and functional analysis of the interaction between the2 nucleoporin nup98 and the mrna export factor rae1
Phyre2
| 16 |
|
PDB 1vtm chain P
Region: 398 - 423
Aligned: 26
Modelled: 26
Confidence: 5.9%
Identity: 19%
Fold: Four-helical up-and-down bundle
Superfamily: TMV-like viral coat proteins
Family: TMV-like viral coat proteins
Phyre2
| 17 |
|
PDB 3c9p chain A
Region: 398 - 409
Aligned: 12
Modelled: 12
Confidence: 5.6%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sp1917;
PDBTitle: crystal structure of uncharacterized protein sp1917
Phyre2
| 18 |
|
PDB 1r9p chain A
Region: 385 - 425
Aligned: 41
Modelled: 41
Confidence: 5.2%
Identity: 17%
Fold: SufE/NifU
Superfamily: SufE/NifU
Family: NifU/IscU domain
Phyre2