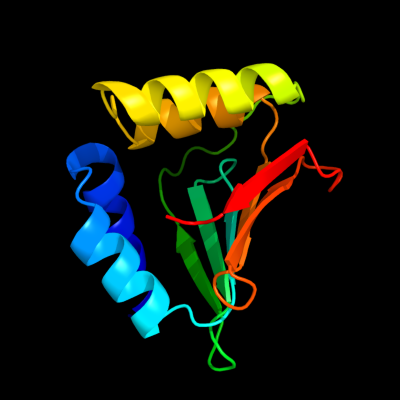
| 1 | d2hh8a1
|
|
 |
100.0 |
80 |
Fold:YdfO-like
Superfamily:YdfO-like
Family:YdfO-like |
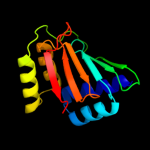
| 2 | d3elga1
|
|
 |
81.9 |
11 |
Fold:BLIP-like
Superfamily:BT0923-like
Family:BT0923-like |
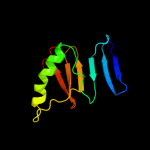
| 3 | c2qlxA_
|
|
 |
41.0 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:l-rhamnose mutarotase;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum in complex with l-rhamnose
|
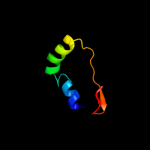
| 4 | c2qlwA_
|
|
 |
41.0 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:rhau;
PDBTitle: crystal structure of rhamnose mutarotase rhau of rhizobium2 leguminosarum
|
| 5 | d3duea1
|
|
 |
39.4 |
4 |
Fold:BLIP-like
Superfamily:BT0923-like
Family:BT0923-like |
| 6 | c1f8aB_
|
|
 |
26.7 |
10 |
PDB header:isomerase
Chain: B: PDB Molecule:peptidyl-prolyl cis-trans isomerase nima-
PDBTitle: structural basis for the phosphoserine-proline recognition2 by group iv ww domains
|
| 7 | c2ph7B_
|
|
 |
26.6 |
38 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein af_2093;
PDBTitle: crystal structure of af2093 from archaeoglobus fulgidus
|
| 8 | c1irjG_
|
|
 |
25.8 |
15 |
PDB header:metal binding protein
Chain: G: PDB Molecule:migration inhibitory factor-related protein 14;
PDBTitle: crystal structure of the mrp14 complexed with chaps
|
| 9 | c2w3nA_
|
|
 |
25.2 |
24 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase 2;
PDBTitle: structure and inhibition of the co2-sensing carbonic2 anhydrase can2 from the pathogenic fungus cryptococcus3 neoformans
|
| 10 | d1x8da1
|
|
 |
24.3 |
14 |
Fold:Ferredoxin-like
Superfamily:Dimeric alpha+beta barrel
Family:YiiL-like |
| 11 | d1avsa_
|
|
 |
23.4 |
17 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 12 | c2k7bA_
|
|
 |
23.2 |
14 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcium-binding protein 1;
PDBTitle: nmr structure of mg2+-bound cabp1 n-domain
|
| 13 | c3l23A_
|
|
 |
21.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of sugar phosphate isomerase/epimerase2 (yp_001303399.1) from parabacteroides distasonis atcc 8503 at 1.70 a3 resolution
|
| 14 | c3ec8A_
|
|
 |
18.7 |
11 |
PDB header:cell adhesion
Chain: A: PDB Molecule:putative uncharacterized protein flj10324;
PDBTitle: the crystal structure of the ra domain of flj10324 (radil)
|
| 15 | c3obeB_
|
|
 |
17.9 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
| 16 | d1xk4a1
|
|
 |
17.4 |
13 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 17 | d1ekja_
|
|
 |
16.6 |
10 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 18 | d1m39a_
|
|
 |
16.4 |
20 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 19 | d1g5ca_
|
|
 |
15.8 |
13 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 20 | c2i18A_
|
|
 |
15.7 |
9 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcium-binding protein;
PDBTitle: the refined structure of c-terminal domain of an ef-hand2 calcium binding protein from entamoeba histolytica
|
| 21 | c3ot2B_ |
|
not modelled |
14.2 |
15 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative nuclease belonging to duf8202 (ava_3926) from anabaena variabilis atcc 29413 at 1.96 a resolution
|
| 22 | c3ot2A_ |
|
not modelled |
14.2 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a putative nuclease belonging to duf8202 (ava_3926) from anabaena variabilis atcc 29413 at 1.96 a resolution
|
| 23 | d1k8ua_ |
|
not modelled |
14.0 |
31 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 24 | d1g33a_ |
|
not modelled |
13.2 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Parvalbumin |
| 25 | d1j55a_ |
|
not modelled |
12.9 |
12 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 26 | c3ucoB_ |
|
not modelled |
12.6 |
15 |
PDB header:lyase/lyase inhibitor
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: coccomyxa beta-carbonic anhydrase in complex with iodide
|
| 27 | c2kaxA_ |
|
not modelled |
12.3 |
23 |
PDB header:metal binding protein
Chain: A: PDB Molecule:protein s100-a5;
PDBTitle: solution structure and dynamics of s100a5 in the apo and2 ca2+ -bound states
|
| 28 | d1e8aa_ |
|
not modelled |
12.2 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 29 | c2rqsA_ |
|
not modelled |
12.2 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:parvulin-like peptidyl-prolyl isomerase;
PDBTitle: 3d structure of pin from the psychrophilic archeon cenarcheaum2 symbiosum (cspin)
|
| 30 | d1pvaa_ |
|
not modelled |
11.7 |
12 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Parvalbumin |
| 31 | d4icba_ |
|
not modelled |
11.6 |
15 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calbindin D9K |
| 32 | d1wxaa1 |
|
not modelled |
11.5 |
6 |
Fold:beta-Grasp (ubiquitin-like)
Superfamily:Ubiquitin-like
Family:Ras-binding domain, RBD |
| 33 | d1i6pa_ |
|
not modelled |
11.2 |
14 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
| 34 | c2rgiA_ |
|
not modelled |
11.1 |
27 |
PDB header:metal binding protein
Chain: A: PDB Molecule:protein s100-a2;
PDBTitle: crystal structure of ca2+-free s100a2 at 1.6 a resolution
|
| 35 | c2hklB_ |
|
not modelled |
11.1 |
9 |
PDB header:transferase
Chain: B: PDB Molecule:l,d-transpeptidase;
PDBTitle: crystal structure of enterococcus faecium l,d-2 transpeptidase c442s mutant
|
| 36 | c2z4hB_ |
|
not modelled |
11.1 |
12 |
PDB header:signaling protein activator
Chain: B: PDB Molecule:copper homeostasis protein cutf;
PDBTitle: crystal structure of the cpx pathway activator nlpe from2 escherichia coli
|
| 37 | c2ggmA_ |
|
not modelled |
11.0 |
14 |
PDB header:cell cycle
Chain: A: PDB Molecule:centrin-2;
PDBTitle: human centrin 2 xeroderma pigmentosum group c protein2 complex
|
| 38 | d1xk4c1 |
|
not modelled |
10.8 |
15 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 39 | c2a4jA_ |
|
not modelled |
10.6 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:centrin 2;
PDBTitle: solution structure of the c-terminal domain (t94-y172) of2 the human centrin 2 in complex with a 17 residues peptide3 (p1-xpc) from xeroderma pigmentosum group c protein
|
| 40 | c2k2aA_ |
|
not modelled |
10.4 |
23 |
PDB header:contractile protein
Chain: A: PDB Molecule:troponin c;
PDBTitle: solution structure of the apo c terminal domain of lethocerus troponin2 c isoform f1
|
| 41 | c2b1uA_ |
|
not modelled |
10.3 |
11 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calmodulin-like protein 5;
PDBTitle: solution structure of calmodulin-like skin protein c2 terminal domain
|
| 42 | c2pmyB_ |
|
not modelled |
10.2 |
24 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:ras and ef-hand domain-containing protein;
PDBTitle: ef-hand domain of human rasef
|
| 43 | c2a5vB_ |
|
not modelled |
10.1 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase (carbonate dehydratase) (carbonic
PDBTitle: crystal structure of m. tuberculosis beta carbonic anhydrase, rv3588c,2 tetrameric form
|
| 44 | d1eq3a_ |
|
not modelled |
9.9 |
5 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 45 | c2a8cE_ |
|
not modelled |
9.9 |
11 |
PDB header:lyase
Chain: E: PDB Molecule:carbonic anhydrase 2;
PDBTitle: haemophilus influenzae beta-carbonic anhydrase
|
| 46 | c1ponB_ |
|
not modelled |
9.7 |
20 |
PDB header:calcium-binding protein
Chain: B: PDB Molecule:troponin c;
PDBTitle: site iii-site iv troponin c heterodimer, nmr
|
| 47 | d1c7va_ |
|
not modelled |
9.6 |
9 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 48 | c3p6lA_ |
|
not modelled |
9.5 |
4 |
PDB header:isomerase
Chain: A: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
|
| 49 | c2kz2A_ |
|
not modelled |
9.4 |
0 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calmodulin;
PDBTitle: calmodulin, c-terminal domain, f92e mutant
|
| 50 | d1oqpa_ |
|
not modelled |
9.4 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 51 | d1zmba1 |
|
not modelled |
9.4 |
12 |
Fold:Flavodoxin-like
Superfamily:SGNH hydrolase
Family:Putative acetylxylan esterase-like |
| 52 | d1a4pa_ |
|
not modelled |
9.3 |
16 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 53 | d1dj0a_ |
|
not modelled |
9.3 |
12 |
Fold:Pseudouridine synthase
Superfamily:Pseudouridine synthase
Family:Pseudouridine synthase I TruA |
| 54 | d1urra_ |
|
not modelled |
9.3 |
21 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 55 | c2l0vB_ |
|
not modelled |
9.2 |
15 |
PDB header:metal binding protein
Chain: B: PDB Molecule:protein s100-a16;
PDBTitle: solution structure of calcium(ii) bound s100a16
|
| 56 | d1apsa_ |
|
not modelled |
8.9 |
7 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 57 | d2p5zx2 |
|
not modelled |
8.8 |
12 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
| 58 | c1q88B_ |
|
not modelled |
8.8 |
11 |
PDB header:dna binding protein
Chain: B: PDB Molecule:39 kda initiator binding protein;
PDBTitle: crystal structure of the c-domain of the t.vaginalis inr2 binding protein, ibp39 (monoclinic form)
|
| 59 | d1ksoa_ |
|
not modelled |
8.6 |
27 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 60 | c3h4sE_ |
|
not modelled |
8.6 |
3 |
PDB header:motor protein/calcium binding protein
Chain: E: PDB Molecule:kcbp interacting ca2+-binding protein;
PDBTitle: structure of the complex of a mitotic kinesin with its2 calcium binding regulator
|
| 61 | d2gaxa1 |
|
not modelled |
8.6 |
19 |
Fold:Peptidyl-tRNA hydrolase II
Superfamily:Peptidyl-tRNA hydrolase II
Family:Peptidyl-tRNA hydrolase II |
| 62 | d1yuta1 |
|
not modelled |
8.2 |
16 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 63 | d2pv2a1 |
|
not modelled |
8.2 |
17 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 64 | d1j6ya_ |
|
not modelled |
8.1 |
9 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 65 | c3ddcB_ |
|
not modelled |
8.1 |
13 |
PDB header:hydrolase/apoptosis
Chain: B: PDB Molecule:ras association domain-containing family protein 5;
PDBTitle: crystal structure of nore1a in complex with ras
|
| 66 | d3d37a1 |
|
not modelled |
8.1 |
17 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:Baseplate protein-like |
| 67 | d2pq3a1 |
|
not modelled |
8.1 |
13 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 68 | d1m45a_ |
|
not modelled |
8.0 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 69 | c2p5zX_ |
|
not modelled |
8.0 |
12 |
PDB header:structural genomics, unknown function
Chain: X: PDB Molecule:type vi secretion system component;
PDBTitle: the e. coli c3393 protein is a component of the type vi secretion2 system and exhibits structural similarity to t4 bacteriophage tail3 proteins gp27 and gp5
|
| 70 | d1cb1a_ |
|
not modelled |
8.0 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calbindin D9K |
| 71 | d1qlka_ |
|
not modelled |
7.9 |
15 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 72 | c1bomB_ |
|
not modelled |
7.8 |
17 |
PDB header:insulin-like brain-secretory peptide
Chain: B: PDB Molecule:bombyxin-ii,bombyxin a-6;
PDBTitle: three-dimensional structure of bombyxin-ii, an insulin-2 related brain-secretory peptide of the silkmoth bombyx3 mori: comparison with insulin and relaxin
|
| 73 | d1wrka1 |
|
not modelled |
7.7 |
10 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 74 | d3cr5x1 |
|
not modelled |
7.7 |
19 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 75 | c3ox6C_ |
|
not modelled |
7.6 |
15 |
PDB header:calcium binding protein
Chain: C: PDB Molecule:calcium-binding protein 1;
PDBTitle: crystal structure of the calcium sensor calcium-binding protein 12 (cabp1)
|
| 76 | d1j3ea_ |
|
not modelled |
7.6 |
24 |
Fold:Replication modulator SeqA, C-terminal DNA-binding domain
Superfamily:Replication modulator SeqA, C-terminal DNA-binding domain
Family:Replication modulator SeqA, C-terminal DNA-binding domain |
| 77 | d1psra_ |
|
not modelled |
7.5 |
17 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:S100 proteins |
| 78 | c3fs7D_ |
|
not modelled |
7.4 |
15 |
PDB header:metal binding protein
Chain: D: PDB Molecule:parvalbumin, thymic;
PDBTitle: crystal structure of gallus gallus beta-parvalbumin (avian2 thymic hormone)
|
| 79 | c3cnyA_ |
|
not modelled |
7.4 |
17 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:inositol catabolism protein iole;
PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution
|
| 80 | d1qx2a_ |
|
not modelled |
7.4 |
13 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calbindin D9K |
| 81 | c2amiA_ |
|
not modelled |
7.2 |
16 |
PDB header:cell cycle
Chain: A: PDB Molecule:caltractin;
PDBTitle: solution structure of the calcium-loaded n-terminal sensor2 domain of centrin
|
| 82 | c2ktgA_ |
|
not modelled |
7.2 |
17 |
PDB header:ca-binding protein
Chain: A: PDB Molecule:calmodulin, putative;
PDBTitle: calmodulin like protein from entamoeba histolytica: solution structure2 and calcium binding properties of a partially folded protein
|
| 83 | d2obha1 |
|
not modelled |
7.2 |
15 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 84 | d1zaca_ |
|
not modelled |
7.1 |
16 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Calmodulin-like |
| 85 | c3lmzA_ |
|
not modelled |
7.0 |
9 |
PDB header:isomerase
Chain: A: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase. (yp_001305105.1) from2 parabacteroides distasonis atcc 8503 at 1.44 a resolution
|
| 86 | d1q88a_ |
|
not modelled |
6.9 |
13 |
Fold:39 kda initiator binding protein, IBP39, C-terminal domains
Superfamily:39 kda initiator binding protein, IBP39, C-terminal domains
Family:39 kda initiator binding protein, IBP39, C-terminal domains |
| 87 | d1ftpa_ |
|
not modelled |
6.9 |
7 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Fatty acid binding protein-like |
| 88 | c2vtgA_ |
|
not modelled |
6.9 |
7 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:ionized calcium-binding adapter molecule 2;
PDBTitle: crystal structure of human iba2, trigonal crystal form
|
| 89 | d2acya_ |
|
not modelled |
6.8 |
10 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 90 | d1m5ya3 |
|
not modelled |
6.8 |
9 |
Fold:FKBP-like
Superfamily:FKBP-like
Family:FKBP immunophilin/proline isomerase |
| 91 | c3br8A_ |
|
not modelled |
6.8 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable acylphosphatase;
PDBTitle: crystal structure of acylphosphatase from bacillus subtilis
|
| 92 | c3ngfA_ |
|
not modelled |
6.7 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:ap endonuclease, family 2;
PDBTitle: crystal structure of ap endonuclease, family 2 from brucella2 melitensis
|
| 93 | c2kdhA_ |
|
not modelled |
6.6 |
11 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: the solution structure of human cardiac troponin c in2 complex with the green tea polyphenol; (-)-3 epigallocatechin-3-gallate
|
| 94 | d1w2ia_ |
|
not modelled |
6.6 |
21 |
Fold:Ferredoxin-like
Superfamily:Acylphosphatase/BLUF domain-like
Family:Acylphosphatase-like |
| 95 | d5pala_ |
|
not modelled |
6.5 |
15 |
Fold:EF Hand-like
Superfamily:EF-hand
Family:Parvalbumin |
| 96 | c1yw5A_ |
|
not modelled |
6.5 |
8 |
PDB header:isomerase
Chain: A: PDB Molecule:peptidyl prolyl cis/trans isomerase;
PDBTitle: peptidyl-prolyl isomerase ess1 from candida albicans
|
| 97 | c2dk6A_ |
|
not modelled |
6.5 |
24 |
PDB header:signaling protein
Chain: A: PDB Molecule:parp11 protein;
PDBTitle: solution structure of wwe domain in poly (adp-ribose)2 polymerase family, member 11 (parp 11)
|
| 98 | c3cu2A_ |
|
not modelled |
6.4 |
12 |
PDB header:isomerase
Chain: A: PDB Molecule:ribulose-5-phosphate 3-epimerase;
PDBTitle: crystal structure of ribulose-5-phosphate 3-epimerase (yp_718263.1)2 from haemophilus somnus 129pt at 1.91 a resolution
|
| 99 | c1ozsA_ |
|
not modelled |
6.4 |
9 |
PDB header:structural protein
Chain: A: PDB Molecule:troponin c, slow skeletal and cardiac muscles;
PDBTitle: c-domain of human cardiac troponin c in complex with the2 inhibitory region of human cardiac troponin i
|