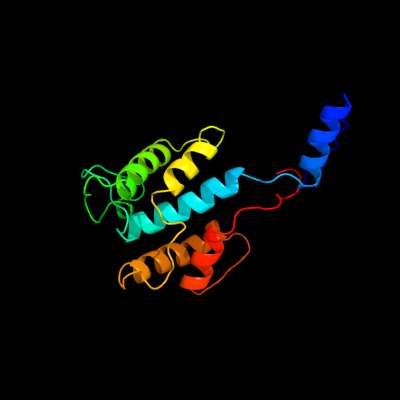
| 1 | d2ijra1
|
|
 |
100.0 |
31 |
Fold:Api92-like
Superfamily:Api92-like
Family:Api92-like |
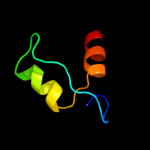
| 2 | d1xs8a_
|
|
 |
58.2 |
21 |
Fold:YggX-like
Superfamily:YggX-like
Family:YggX-like |
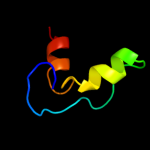
| 3 | d1t07a_
|
|
 |
54.8 |
21 |
Fold:YggX-like
Superfamily:YggX-like
Family:YggX-like |
| 4 | c1ec8B_
|
|
 |
37.6 |
31 |
PDB header:lyase
Chain: B: PDB Molecule:glucarate dehydratase;
PDBTitle: e. coli glucarate dehydratase bound to product 2,3-2 dihydroxy-5-oxo-hexanedioate
|
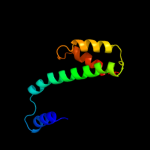
| 5 | c1tndA_
|
|
 |
26.8 |
16 |
PDB header:binding protein(gtp)
Chain: A: PDB Molecule:transducin;
PDBTitle: the 2.2 angstroms crystal structure of transducin-alpha complexed with2 gtp gamma s
|
| 6 | c2xtzB_
|
|
 |
22.8 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:guanine nucleotide-binding protein alpha-1 subunit;
PDBTitle: crystal structure of the g alpha protein atgpa1 from2 arabidopsis thaliana
|
| 7 | d1saza1
|
|
 |
22.2 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Acetokinase-like |
| 8 | d2bj7a1
|
|
 |
20.4 |
13 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 9 | c2ca9B_
|
|
 |
20.4 |
13 |
PDB header:transcriptional regulation
Chain: B: PDB Molecule:putative nickel-responsive regulator;
PDBTitle: apo-nikr from helicobacter pylori in closed trans-2 conformation
|
| 10 | c1zcbA_
|
|
 |
18.4 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:g alpha i/13;
PDBTitle: crystal structure of g alpha 13 in complex with gdp
|
| 11 | c3n6jA_
|
|
 |
16.8 |
28 |
PDB header:isomerase
Chain: A: PDB Molecule:mandelate racemase/muconate lactonizing protein;
PDBTitle: crystal structure of mandelate racemase/muconate lactonizing protein2 from actinobacillus succinogenes 130z
|
| 12 | c1tl7C_
|
|
 |
14.5 |
23 |
PDB header:lyase
Chain: C: PDB Molecule:guanine nucleotide-binding protein g(s), alpha
PDBTitle: complex of gs- with the catalytic domains of mammalian2 adenylyl cyclase: complex with 2'(3')-o-(n-3 methylanthraniloyl)-guanosine 5'-triphosphate and mn
|
| 13 | c3ijmA_
|
|
 |
13.0 |
18 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized restriction endonuclease-like fold
PDBTitle: the structure of a restriction endonuclease-like fold superfamily2 protein from spirosoma linguale.
|
| 14 | d2ihoa2
|
|
 |
12.9 |
26 |
Fold:Cysteine proteinases
Superfamily:Cysteine proteinases
Family:MOA C-terminal domain-like |
| 15 | d2a90a2
|
|
 |
12.2 |
18 |
Fold:WWE domain
Superfamily:WWE domain
Family:WWE domain |
| 16 | c1cipA_
|
|
 |
12.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (guanine nucleotide-binding protein
PDBTitle: gi-alpha-1 subunit of guanine nucleotide-binding protein2 complexed with a gtp analogue
|
| 17 | c3ue9A_
|
|
 |
12.1 |
36 |
PDB header:ligase
Chain: A: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: crystal structure of adenylosuccinate synthetase (ampsase) (pura) from2 burkholderia thailandensis
|
| 18 | d1osce_
|
|
 |
11.8 |
23 |
Fold:Ferredoxin-like
Superfamily:GlnB-like
Family:Divalent ion tolerance proteins CutA (CutA1) |
| 19 | d1iwea_
|
|
 |
11.6 |
36 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 20 | c1iweB_
|
|
 |
11.4 |
36 |
PDB header:ligase
Chain: B: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: imp complex of the recombinant mouse-muscle2 adenylosuccinate synthetase
|
| 21 | d1ngka_ |
|
not modelled |
10.6 |
16 |
Fold:Globin-like
Superfamily:Globin-like
Family:Truncated hemoglobin |
| 22 | c3cx6A_ |
|
not modelled |
10.3 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:guanine nucleotide-binding protein alpha-13
PDBTitle: crystal structure of pdzrhogef rgrgs domain in a complex2 with galpha-13 bound to gdp
|
| 23 | d2ozka1 |
|
not modelled |
10.0 |
56 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Nsp15 N-terminal domain-like |
| 24 | c2bcjQ_ |
|
not modelled |
9.9 |
12 |
PDB header:transferase/hydrolase
Chain: Q: PDB Molecule:g alpha i1, guanine nucleotide-binding protein g(q), alpha
PDBTitle: crystal structure of g protein-coupled receptor kinase 2 in complex2 with galpha-q and gbetagamma subunits
|
| 25 | d2guka1 |
|
not modelled |
9.5 |
27 |
Fold:PG1857-like
Superfamily:PG1857-like
Family:PG1857-like |
| 26 | c3iuzA_ |
|
not modelled |
9.1 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:putative glyoxalase superfamily protein;
PDBTitle: crystal structure of putative glyoxalase superfamily protein2 (yp_299723.1) from ralstonia eutropha jmp134 at 1.90 a resolution
|
| 27 | d2gqfa2 |
|
not modelled |
8.9 |
3 |
Fold:HI0933 insert domain-like
Superfamily:HI0933 insert domain-like
Family:HI0933 insert domain-like |
| 28 | d1jdfa1 |
|
not modelled |
8.9 |
38 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
| 29 | d2h85a1 |
|
not modelled |
8.6 |
56 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Nsp15 N-terminal domain-like |
| 30 | c1sxjB_ |
|
not modelled |
8.1 |
22 |
PDB header:replication
Chain: B: PDB Molecule:activator 1 37 kda subunit;
PDBTitle: crystal structure of the eukaryotic clamp loader2 (replication factor c, rfc) bound to the dna sliding clamp3 (proliferating cell nuclear antigen, pcna)
|
| 31 | d1p9ba_ |
|
not modelled |
8.1 |
28 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 32 | d1ac5a_ |
|
not modelled |
8.0 |
13 |
Fold:alpha/beta-Hydrolases
Superfamily:alpha/beta-Hydrolases
Family:Serine carboxypeptidase-like |
| 33 | d1sxjc2 |
|
not modelled |
7.9 |
19 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 34 | c3thgA_ |
|
not modelled |
7.8 |
14 |
PDB header:protein binding
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase activase 1,
PDBTitle: crystal structure of the creosote rubisco activase c-domain
|
| 35 | d2bcjq1 |
|
not modelled |
7.6 |
11 |
Fold:Transducin (alpha subunit), insertion domain
Superfamily:Transducin (alpha subunit), insertion domain
Family:Transducin (alpha subunit), insertion domain |
| 36 | c2d7uA_ |
|
not modelled |
7.6 |
24 |
PDB header:ligase
Chain: A: PDB Molecule:adenylosuccinate synthetase;
PDBTitle: crystal structure of hypothetical adenylosuccinate synthetase, ph04382 from pyrococcus horikoshii ot3
|
| 37 | c1iqpF_ |
|
not modelled |
7.5 |
25 |
PDB header:replication
Chain: F: PDB Molecule:rfcs;
PDBTitle: crystal structure of the clamp loader small subunit from2 pyrococcus furiosus
|
| 38 | c3gnzP_ |
|
not modelled |
7.3 |
25 |
PDB header:toxin
Chain: P: PDB Molecule:25 kda protein elicitor;
PDBTitle: toxin fold for microbial attack and plant defense
|
| 39 | c2zzxD_ |
|
not modelled |
7.3 |
32 |
PDB header:transport protein
Chain: D: PDB Molecule:abc transporter, solute-binding protein;
PDBTitle: crystal structure of a periplasmic substrate binding protein in2 complex with lactate
|
| 40 | c1rrzA_ |
|
not modelled |
7.2 |
33 |
PDB header:structural genomics,biosynthetic protein
Chain: A: PDB Molecule:glycogen synthesis protein glgs;
PDBTitle: solution structure of glgs protein from e. coli
|
| 41 | d1rrza_ |
|
not modelled |
7.2 |
33 |
Fold:Spectrin repeat-like
Superfamily:Glycogen synthesis protein GlgS
Family:Glycogen synthesis protein GlgS |
| 42 | c2xykB_ |
|
not modelled |
7.0 |
13 |
PDB header:oxygen storage/transport
Chain: B: PDB Molecule:2-on-2 hemoglobin;
PDBTitle: group ii 2-on-2 hemoglobin from the plant pathogen2 agrobacterium tumefaciens
|
| 43 | d2ixma1 |
|
not modelled |
7.0 |
19 |
Fold:PTPA-like
Superfamily:PTPA-like
Family:PTPA-like |
| 44 | d1zcaa1 |
|
not modelled |
6.8 |
23 |
Fold:Transducin (alpha subunit), insertion domain
Superfamily:Transducin (alpha subunit), insertion domain
Family:Transducin (alpha subunit), insertion domain |
| 45 | d1dj2a_ |
|
not modelled |
6.6 |
20 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Nitrogenase iron protein-like |
| 46 | d2hzaa1 |
|
not modelled |
6.5 |
13 |
Fold:Ribbon-helix-helix
Superfamily:Ribbon-helix-helix
Family:CopG-like |
| 47 | d1nmla2 |
|
not modelled |
5.8 |
22 |
Fold:Cytochrome c
Superfamily:Cytochrome c
Family:Di-heme cytochrome c peroxidase |
| 48 | d2ixoa1 |
|
not modelled |
5.8 |
25 |
Fold:PTPA-like
Superfamily:PTPA-like
Family:PTPA-like |
| 49 | c2bj3D_ |
|
not modelled |
5.5 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:nickel responsive regulator;
PDBTitle: nikr-apo
|
| 50 | c2chgB_ |
|
not modelled |
5.5 |
22 |
PDB header:dna-binding protein
Chain: B: PDB Molecule:replication factor c small subunit;
PDBTitle: replication factor c domains 1 and 2
|
| 51 | d1pyla_ |
|
not modelled |
5.3 |
12 |
Fold:Microbial ribonucleases
Superfamily:Microbial ribonucleases
Family:Bacterial ribonucleases |
| 52 | c2a3zC_ |
|
not modelled |
5.2 |
57 |
PDB header:structural protein
Chain: C: PDB Molecule:wiskott-aldrich syndrome protein;
PDBTitle: ternary complex of the wh2 domain of wasp with actin-dnase i
|
| 53 | c3d33B_ |
|
not modelled |
5.2 |
33 |
PDB header:unknown function
Chain: B: PDB Molecule:domain of unknown function with an immunoglobulin-like
PDBTitle: crystal structure of a duf3244 family protein with an immunoglobulin-2 like beta-sandwich fold (bvu_0276) from bacteroides vulgatus atcc3 8482 at 1.70 a resolution
|
| 54 | c2g62A_ |
|
not modelled |
5.2 |
25 |
PDB header:hydrolase activator
Chain: A: PDB Molecule:protein phosphatase 2a, regulatory subunit b' (pr 53);
PDBTitle: crystal structure of human ptpa
|
| 55 | c2rhbD_ |
|
not modelled |
5.1 |
56 |
PDB header:viral protein
Chain: D: PDB Molecule:uridylate-specific endoribonuclease;
PDBTitle: crystal structure of nsp15-h234a mutant- hexamer in2 asymmetric unit
|
| 56 | d1sxjb2 |
|
not modelled |
5.1 |
22 |
Fold:P-loop containing nucleoside triphosphate hydrolases
Superfamily:P-loop containing nucleoside triphosphate hydrolases
Family:Extended AAA-ATPase domain |
| 57 | c2nz7A_ |
|
not modelled |
5.1 |
21 |
PDB header:apoptosis
Chain: A: PDB Molecule:caspase recruitment domain-containing protein 4;
PDBTitle: crystal structure analysis of caspase-recruitment domain2 (card) of nod1
|
| 58 | c1z7s1_ |
|
not modelled |
5.0 |
38 |
PDB header:virus
Chain: 1: PDB Molecule:human coxsackievirus a21;
PDB Fragment:viral protein 1;
PDBTitle: the crystal structure of coxsackievirus a21
|