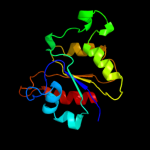
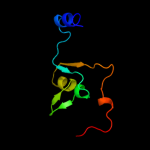
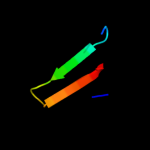
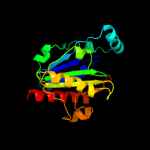
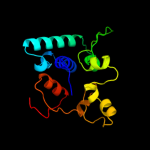
1 c2gp4A_
100.0
28
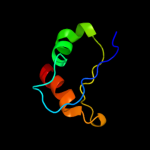
PDB header: lyaseChain: A: PDB Molecule: 6-phosphogluconate dehydratase;PDBTitle: structure of [fes]cluster-free apo form of 6-phosphogluconate2 dehydratase from shewanella oneidensis
2 d2gp4a2
100.0
27
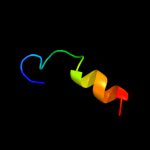
Fold: IlvD/EDD N-terminal domain-likeSuperfamily: IlvD/EDD N-terminal domain-likeFamily: lvD/EDD N-terminal domain-like3 d2gp4a1
100.0
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: IlvD/EDD C-terminal domain-like4 c2pcnA_
79.3
17
PDB header: transferaseChain: A: PDB Molecule: s-adenosylmethionine:2-demethylmenaquinonePDBTitle: crystal structure of s-adenosylmethionine: 2-dimethylmenaquinone2 methyltransferase (gk_1813) from geobacillus kaustophilus hta426
5 c2jv2A_
69.3
25
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein ph1500;PDBTitle: solution structure of the n-terminal domain of ph1500
6 d1nxja_
66.5
15
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like7 c1nxjA_
66.5
15
PDB header: unknown functionChain: A: PDB Molecule: probable s-adenosylmethionine:2-PDBTitle: structure of rv3853 from mycobacterium tuberculosis
8 c3c8oB_
64.9
18
PDB header: hydrolase regulatorChain: B: PDB Molecule: regulator of ribonuclease activity a;PDBTitle: the crystal structure of rraa from pao1
9 d1vi4a_
62.5
13
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like10 d1q5xa_
52.4
15
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like11 d1agxa_
51.7
15
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase12 d1pl8a1
50.0
21
Fold: GroES-likeSuperfamily: GroES-likeFamily: Alcohol dehydrogenase-like, N-terminal domain13 d1wsaa_
45.6
21
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase14 d1y0pa2
42.7
23
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain15 d2hi6a1
40.6
11
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: AF0055-like16 d1ydga_
37.2
13
Fold: Flavodoxin-likeSuperfamily: FlavoproteinsFamily: WrbA-like17 d1fjca_
35.2
34
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD18 d1wl8a1
35.1
23
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)19 d1r31a1
35.0
35
Fold: Ferredoxin-likeSuperfamily: NAD-binding domain of HMG-CoA reductaseFamily: NAD-binding domain of HMG-CoA reductase20 d1j33a_
34.4
19
Fold: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Superfamily: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Family: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)21 d1od5a1
not modelled
34.0
18
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein22 d1v7la_
not modelled
32.1
17
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like23 d1zyma2
not modelled
32.0
9
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: N-terminal domain of enzyme I of the PEP:sugar phosphotransferase system24 c1dm9A_
not modelled
32.0
15
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical 15.5 kd protein in mrca-pckaPDBTitle: heat shock protein 15 kd
25 d1dm9a_
not modelled
32.0
15
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Heat shock protein 15 kD26 d1kola2
not modelled
31.9
24
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain27 d1j3la_
not modelled
31.8
26
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: RraA-likeFamily: RraA-like28 c1sddA_
not modelled
31.5
21
PDB header: blood clottingChain: A: PDB Molecule: coagulation factor v;PDBTitle: crystal structure of bovine factor vai
29 d1ujva_
not modelled
31.4
32
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: PDZ domain30 d1o7ja_
not modelled
31.0
12
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase31 d1r61a_
not modelled
28.7
15
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Putative cyclaseFamily: Putative cyclase32 d1rk8a_
not modelled
28.4
21
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD33 c1rkjA_
not modelled
26.9
30
PDB header: transcription/rnaChain: A: PDB Molecule: nucleolin;PDBTitle: solution structure of the complex formed by the two n-2 terminal rna-binding domains of nucleolin and a pre-rrna3 target
34 c3l83A_
not modelled
25.9
20
PDB header: transferaseChain: A: PDB Molecule: glutamine amido transferase;PDBTitle: crystal structure of glutamine amido transferase from methylobacillus2 flagellatus
35 d1a9xb2
not modelled
25.3
22
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)36 d1lcya1
not modelled
25.0
34
Fold: PDZ domain-likeSuperfamily: PDZ domain-likeFamily: HtrA-like serine proteases37 d2etna2
not modelled
24.4
14
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain38 c2qv5A_
not modelled
24.3
26
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein atu2773;PDBTitle: crystal structure of uncharacterized protein atu2773 from2 agrobacterium tumefaciens c58
39 c3nojA_
not modelled
24.2
29
PDB header: lyaseChain: A: PDB Molecule: 4-carboxy-4-hydroxy-2-oxoadipate aldolase/oxaloacetatePDBTitle: the structure of hmg/cha aldolase from the protocatechuate degradation2 pathway of pseudomonas putida
40 c3t6oA_
not modelled
24.1
18
PDB header: transport proteinChain: A: PDB Molecule: sulfate transporter/antisigma-factor antagonist stas;PDBTitle: the structure of an anti-sigma-factor antagonist (stas) domain protein2 from planctomyces limnophilus.
41 c3f43A_
not modelled
24.0
12
PDB header: transcriptionChain: A: PDB Molecule: putative anti-sigma factor antagonist tm1081;PDBTitle: crystal structure of a putative anti-sigma factor antagonist (tm1081)2 from thermotoga maritima at 1.59 a resolution
42 c3kw8A_
not modelled
23.6
21
PDB header: oxidoreductaseChain: A: PDB Molecule: putative copper oxidase;PDBTitle: two-domain laccase from streptomyces coelicolor at 2.3 a resolution
43 d2et1a1
not modelled
23.3
14
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein44 d1k78a1
not modelled
22.9
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain45 c3fm3B_
not modelled
22.9
19
PDB header: hydrolaseChain: B: PDB Molecule: methionine aminopeptidase 2;PDBTitle: crystal structure of an encephalitozoon cuniculi methionine2 aminopeptidase type 2
46 d2bs2a2
not modelled
22.8
26
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: Succinate dehydrogenase/fumarate reductase flavoprotein N-terminal domain47 c1yw7A_
not modelled
22.4
32
PDB header: hydrolaseChain: A: PDB Molecule: methionine aminopeptidase 2;PDBTitle: h-metap2 complexed with a444148
48 d6paxa1
not modelled
21.5
31
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Paired domain49 d1kbla2
not modelled
21.5
12
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain50 d2f23a2
not modelled
21.5
14
Fold: FKBP-likeSuperfamily: FKBP-likeFamily: GreA transcript cleavage factor, C-terminal domain51 c2pkpA_
not modelled
21.3
24
PDB header: lyaseChain: A: PDB Molecule: homoaconitase small subunit;PDBTitle: crystal structure of 3-isopropylmalate dehydratase (leud)2 from methhanocaldococcus jannaschii dsm2661 (mj1271)
52 d1qdlb_
not modelled
21.1
18
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)53 d1sdda1
not modelled
20.7
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins54 c3ln3A_
not modelled
20.3
14
PDB header: oxidoreductaseChain: A: PDB Molecule: dihydrodiol dehydrogenase;PDBTitle: crystal structure of putative reductase (np_038806.2) from2 mus musculus at 1.18 a resolution
55 c3mvnA_
not modelled
19.9
10
PDB header: ligaseChain: A: PDB Molecule: udp-n-acetylmuramate:l-alanyl-gamma-d-glutamayl-medo-PDBTitle: crystal structure of a domain from a putative udp-n-acetylmuramate:l-2 alanyl-gamma-d-glutamayl-medo-diaminopimelate ligase from haemophilus3 ducreyi 35000hp
56 d1jb0k_
not modelled
18.8
44
Fold: Photosystem I reaction center subunit X, PsaKSuperfamily: Photosystem I reaction center subunit X, PsaKFamily: Photosystem I reaction center subunit X, PsaK57 c1jb0K_
not modelled
18.8
44
PDB header: photosynthesisChain: K: PDB Molecule: photosystem 1 reaction centre subunit x;PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
58 c1ezaA_
not modelled
18.8
14
PDB header: phosphotransferaseChain: A: PDB Molecule: enzyme i;PDBTitle: amino terminal domain of enzyme i from escherichia coli nmr,2 restrained regularized mean structure
59 c2cdqB_
not modelled
18.0
26
PDB header: transferaseChain: B: PDB Molecule: aspartokinase;PDBTitle: crystal structure of arabidopsis thaliana aspartate kinase2 complexed with lysine and s-adenosylmethionine
60 d1vbga2
not modelled
17.8
12
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain61 d1ueba3
not modelled
17.8
6
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like62 c1wa1X_
not modelled
17.7
16
PDB header: reductaseChain: X: PDB Molecule: dissimilatory copper-containing nitritePDBTitle: crystal structure of h313q mutant of alcaligenes2 xylosoxidans nitrite reductase
63 c2hcuA_
not modelled
17.6
11
PDB header: lyaseChain: A: PDB Molecule: 3-isopropylmalate dehydratase small subunit;PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus2 mutans
64 c3cdzA_
not modelled
17.2
21
PDB header: blood clottingChain: A: PDB Molecule: coagulation factor viii heavy chain;PDBTitle: crystal structure of human factor viii
65 c2y0fD_
not modelled
17.1
20
PDB header: oxidoreductaseChain: D: PDB Molecule: 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;PDBTitle: structure of gcpe (ispg) from thermus thermophilus hb27
66 d2cqaa1
not modelled
17.1
18
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: TIP49 domain67 c3soeA_
not modelled
17.1
22
PDB header: signaling proteinChain: A: PDB Molecule: membrane-associated guanylate kinase, ww and pdz domain-PDBTitle: crystal structure of the 3rd pdz domain of the human membrane-2 associated guanylate kinase, ww and pdz domain-containing protein 33 (magi3)
68 c1gph1_
not modelled
17.0
22
PDB header: transferase(glutamine amidotransferase)Chain: 1: PDB Molecule: glutamine phosphoribosyl-pyrophosphate amidotransferase;PDBTitle: structure of the allosteric regulatory enzyme of purine biosynthesis
69 d1l5oa_
not modelled
16.6
18
Fold: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Superfamily: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)Family: Nicotinate mononucleotide:5,6-dimethylbenzimidazole phosphoribosyltransferase (CobT)70 c2p4vA_
not modelled
16.4
10
PDB header: transcriptionChain: A: PDB Molecule: transcription elongation factor greb;PDBTitle: crystal structure of the transcript cleavage factor, greb2 at 2.6a resolution
71 c2e5hA_
not modelled
16.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: zinc finger cchc-type and rna-binding motif-PDBTitle: solution structure of rna binding domain in zinc finger2 cchc-type and rna binding motif 1
72 c2c5qE_
not modelled
16.0
8
PDB header: structural genomics,unknown functionChain: E: PDB Molecule: rraa-like protein yer010c;PDBTitle: crystal structure of yeast yer010cp
73 d1p3da2
not modelled
16.0
19
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain74 c1ecjB_
not modelled
15.7
16
PDB header: transferaseChain: B: PDB Molecule: glutamine phosphoribosylpyrophosphatePDBTitle: escherichia coli glutamine phosphoribosylpyrophosphate2 (prpp) amidotransferase complexed with 2 amp per tetramer
75 d2j5wa4
not modelled
15.7
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins76 c3lzkC_
not modelled
15.4
25
PDB header: hydrolaseChain: C: PDB Molecule: fumarylacetoacetate hydrolase family protein;PDBTitle: the crystal structure of a probably aromatic amino acid2 degradation protein from sinorhizobium meliloti 1021
77 c3nyeA_
not modelled
15.2
30
PDB header: oxidoreductaseChain: A: PDB Molecule: d-arginine dehydrogenase;PDBTitle: crystal structure of pseudomonas aeruginosa d-arginine dehydrogenase2 in complex with imino-arginine
78 d1kv7a1
not modelled
15.2
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins79 d1oe1a1
not modelled
15.1
16
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins80 d2a9va1
not modelled
15.0
23
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)81 c1kzlA_
not modelled
15.0
23
PDB header: transferaseChain: A: PDB Molecule: riboflavin synthase;PDBTitle: riboflavin synthase from s.pombe bound to2 carboxyethyllumazine
82 d1lvla1
not modelled
14.9
32
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains83 d1l5ja2
not modelled
14.7
18
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: LeuD/IlvD-likeFamily: LeuD-like84 c2zooA_
not modelled
14.5
26
PDB header: oxidoreductaseChain: A: PDB Molecule: probable nitrite reductase;PDBTitle: crystal structure of nitrite reductase from pseudoalteromonas2 haloplanktis tac125
85 d2j5wa3
not modelled
14.5
21
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins86 d1e3ja2
not modelled
14.1
36
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain87 c3u7jA_
not modelled
13.9
20
PDB header: isomeraseChain: A: PDB Molecule: ribose-5-phosphate isomerase a;PDBTitle: crystal structure of ribose-5-phosphate isomerase a from burkholderia2 thailandensis
88 d1h6za2
not modelled
13.9
24
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: Phosphohistidine domainFamily: Pyruvate phosphate dikinase, central domain89 c3g5wC_
not modelled
13.7
16
PDB header: metal binding proteinChain: C: PDB Molecule: multicopper oxidase type 1;PDBTitle: crystal structure of blue copper oxidase from nitrosomonas europaea
90 d1e8ca2
not modelled
13.7
24
Fold: MurD-like peptide ligases, peptide-binding domainSuperfamily: MurD-like peptide ligases, peptide-binding domainFamily: MurCDEF C-terminal domain91 d1o1ya_
not modelled
13.6
25
Fold: Flavodoxin-likeSuperfamily: Class I glutamine amidotransferase-likeFamily: Class I glutamine amidotransferases (GAT)92 d1fxza2
not modelled
13.4
13
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein93 c2e28A_
not modelled
13.4
20
PDB header: transferaseChain: A: PDB Molecule: pyruvate kinase;PDBTitle: crystal structure analysis of pyruvate kinase from bacillus2 stearothermophilus
94 d1pl8a2
not modelled
13.2
35
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain95 d1aoza1
not modelled
13.1
16
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Multidomain cupredoxins96 d1ykfa2
not modelled
13.1
27
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Alcohol dehydrogenase-like, C-terminal domain97 c3cgnA_
not modelled
13.0
16
PDB header: isomeraseChain: A: PDB Molecule: peptidyl-prolyl cis-trans isomerase;PDBTitle: crystal structure of thermophilic slyd
98 c1yq4A_
not modelled
13.0
24
PDB header: oxidoreductaseChain: A: PDB Molecule: succinate dehydrogenase flavoprotein subunit;PDBTitle: avian respiratory complex ii with 3-nitropropionate and ubiquinone
99 c3fijD_
not modelled
12.9
18
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: lin1909 protein;PDBTitle: crystal structure of a uncharacterized protein lin1909