| 1 |
|
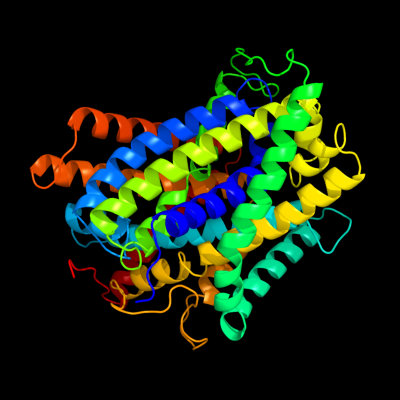
PDB 3gia chain A
Region: 7 - 442
Aligned: 424
Modelled: 431
Confidence: 100.0%
Identity: 19%
PDB header:transport protein
Chain: A: PDB Molecule:uncharacterized protein mj0609;
PDBTitle: crystal structure of apct transporter
Phyre2
| 2 |
|
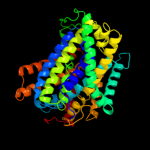
PDB 3lrc chain C
Region: 7 - 442
Aligned: 402
Modelled: 402
Confidence: 100.0%
Identity: 23%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 3 |
|
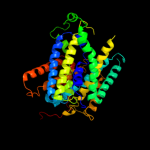
PDB 2jln chain A
Region: 3 - 443
Aligned: 431
Modelled: 431
Confidence: 100.0%
Identity: 11%
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
Phyre2
| 4 |
|
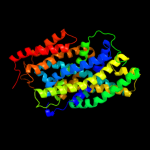
PDB 2xq2 chain A
Region: 8 - 443
Aligned: 427
Modelled: 432
Confidence: 99.6%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: structure of the k294a mutant of vsglt
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 8 - 443
Aligned: 429
Modelled: 429
Confidence: 99.5%
Identity: 9%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 2a65 chain A domain 1
Region: 11 - 440
Aligned: 423
Modelled: 423
Confidence: 98.2%
Identity: 10%
Fold: SNF-like
Superfamily: SNF-like
Family: SNF-like
Phyre2
| 7 |
|
PDB 2w8a chain C
Region: 1 - 386
Aligned: 379
Modelled: 386
Confidence: 96.9%
Identity: 12%
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
Phyre2
| 8 |
|
PDB 3hfx chain A
Region: 7 - 382
Aligned: 369
Modelled: 369
Confidence: 49.9%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:l-carnitine/gamma-butyrobetaine antiporter;
PDBTitle: crystal structure of carnitine transporter
Phyre2
| 9 |
|
PDB 2klu chain A
Region: 419 - 443
Aligned: 25
Modelled: 25
Confidence: 28.5%
Identity: 24%
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
Phyre2
| 10 |
|
PDB 3p5n chain A
Region: 389 - 442
Aligned: 54
Modelled: 54
Confidence: 11.0%
Identity: 11%
PDB header:transport protein
Chain: A: PDB Molecule:riboflavin uptake protein;
PDBTitle: structure and mechanism of the s component of a bacterial ecf2 transporter
Phyre2
| 11 |
|
PDB 2gfp chain A
Region: 319 - 443
Aligned: 125
Modelled: 125
Confidence: 7.7%
Identity: 8%
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
Phyre2
| 12 |
|
PDB 2bbj chain B
Region: 392 - 438
Aligned: 47
Modelled: 47
Confidence: 7.6%
Identity: 6%
PDB header:metal transport/membrane protein
Chain: B: PDB Molecule:divalent cation transport-related protein;
PDBTitle: crystal structure of the cora mg2+ transporter
Phyre2
| 13 |
|
PDB 2jwa chain A
Region: 408 - 442
Aligned: 35
Modelled: 35
Confidence: 7.1%
Identity: 11%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2