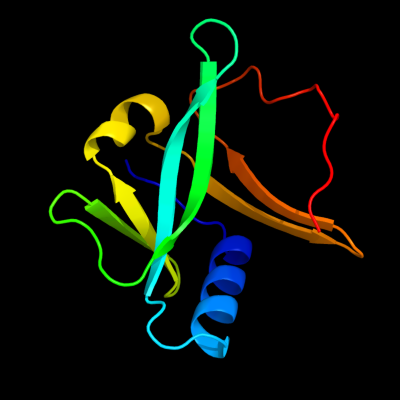
| 1 | c2d2aA_
|
|
 |
100.0 |
47 |
PDB header:metal transport
Chain: A: PDB Molecule:sufa protein;
PDBTitle: crystal structure of escherichia coli sufa involved in2 biosynthesis of iron-sulfur clusters
|
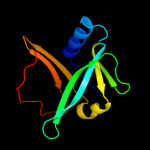
| 2 | c2apnA_
|
|
 |
100.0 |
33 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:protein hi1723;
PDBTitle: hi1723 solution structure
|
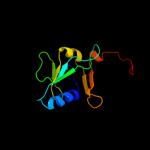
| 3 | c1x0gA_
|
|
 |
100.0 |
31 |
PDB header:metal binding protein
Chain: A: PDB Molecule:isca;
PDBTitle: crystal structure of isca with the [2fe-2s] cluster
|
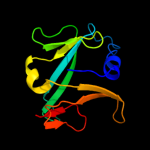
| 4 | d1s98a_
|
|
 |
100.0 |
99 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 5 | c2k4zA_
|
|
 |
100.0 |
31 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:dsrr;
PDBTitle: solution nmr structure of allochromatium vinosum dsrr:2 northeast structural genomics consortium target op5
|
| 6 | d1nwba_
|
|
 |
100.0 |
33 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 7 | d2p2ea1
|
|
 |
98.9 |
19 |
Fold:HesB-like domain
Superfamily:HesB-like domain
Family:HesB-like domain |
| 8 | c2qgoA_
|
|
 |
98.7 |
15 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:putative fe-s biosynthesis protein;
PDBTitle: crystal structure of a putative fe-s biosynthesis protein from2 lactobacillus acidophilus
|
| 9 | d2a6sa1
|
|
 |
19.1 |
10 |
Fold:RelE-like
Superfamily:RelE-like
Family:YoeB/Txe-like |
| 10 | d1sp2a_
|
|
 |
12.6 |
44 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 11 | d1zfda_
|
|
 |
10.6 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 12 | c3kxeD_
|
|
 |
10.5 |
28 |
PDB header:protein binding
Chain: D: PDB Molecule:antitoxin protein pard-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|
| 13 | c3g5oC_
|
|
 |
10.0 |
13 |
PDB header:toxin/antitoxin
Chain: C: PDB Molecule:uncharacterized protein rv2866;
PDBTitle: the crystal structure of the toxin-antitoxin complex relbe2 (rv2865-2 2866) from mycobacterium tuberculosis
|
| 14 | c2yujA_
|
|
 |
9.9 |
33 |
PDB header:protein binding
Chain: A: PDB Molecule:ubiquitin fusion degradation 1-like;
PDBTitle: solution structure of human ubiquitin fusion degradation2 protein 1 homolog ufd1
|
| 15 | c2kdxA_
|
|
 |
9.5 |
22 |
PDB header:metal-binding protein
Chain: A: PDB Molecule:hydrogenase/urease nickel incorporation protein
PDBTitle: solution structure of hypa protein
|
| 16 | d1a1ia1
|
|
 |
9.4 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 17 | d1aaya1
|
|
 |
9.0 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 18 | d1ubdc3
|
|
 |
8.8 |
57 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 19 | c1zc1A_
|
|
 |
8.7 |
23 |
PDB header:protein turnover
Chain: A: PDB Molecule:ubiquitin fusion degradation protein 1;
PDBTitle: ufd1 exhibits the aaa-atpase fold with two distinct2 ubiquitin interaction sites
|
| 20 | c3c3jA_
|
|
 |
8.7 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of tagatose-6-phosphate ketose/aldose isomerase from2 escherichia coli
|
| 21 | d1a1ha1 |
|
not modelled |
8.6 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 22 | d1lk5a2 |
|
not modelled |
8.2 |
24 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 23 | d1a1ga1 |
|
not modelled |
8.2 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 24 | d1f2ig1 |
|
not modelled |
8.1 |
38 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 25 | d1tf3a2 |
|
not modelled |
7.8 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 26 | d1u86a1 |
|
not modelled |
7.8 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 27 | d1wgea1 |
|
not modelled |
7.7 |
40 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 28 | c3odpA_ |
|
not modelled |
7.5 |
38 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tagatose-6-phosphate ketose/aldose isomerase;
PDBTitle: crystal structure of a putative tagatose-6-phosphate ketose/aldose2 isomerase (nt01cx_0292) from clostridium novyi nt at 2.35 a3 resolution
|
| 29 | c3a44D_ |
|
not modelled |
7.5 |
12 |
PDB header:metal binding protein
Chain: D: PDB Molecule:hydrogenase nickel incorporation protein hypa;
PDBTitle: crystal structure of hypa in the dimeric form
|
| 30 | c2kvfA_ |
|
not modelled |
7.3 |
38 |
PDB header:transcription
Chain: A: PDB Molecule:zinc finger and btb domain-containing protein 32;
PDBTitle: structure of the three-cys2his2 domain of mouse testis zinc2 finger protein
|
| 31 | d1adta2 |
|
not modelled |
6.7 |
27 |
Fold:Zn-binding domains of ADDBP
Superfamily:Zn-binding domains of ADDBP
Family:Zn-binding domains of ADDBP |
| 32 | c2wh44_ |
|
not modelled |
6.6 |
80 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: insights into translational termination from the structure2 of rf2 bound to the ribosome
|
| 33 | c1gxsC_ |
|
not modelled |
6.6 |
14 |
PDB header:lyase
Chain: C: PDB Molecule:p-(s)-hydroxymandelonitrile lyase chain a;
PDBTitle: crystal structure of hydroxynitrile lyase from sorghum2 bicolor in complex with inhibitor benzoic acid: a novel3 cyanogenic enzyme
|
| 34 | d1wmia1 |
|
not modelled |
6.4 |
0 |
Fold:RelE-like
Superfamily:RelE-like
Family:RelE-like |
| 35 | c2ab3A_ |
|
not modelled |
6.2 |
67 |
PDB header:rna binding protein
Chain: A: PDB Molecule:znf29;
PDBTitle: solution structures and characterization of hiv rre iib rna2 targeting zinc finger proteins
|
| 36 | c3f1f4_ |
|
not modelled |
6.0 |
80 |
PDB header:ribosome
Chain: 4: PDB Molecule:50s ribosomal protein l31;
PDBTitle: crystal structure of a translation termination complex2 formed with release factor rf2. this file contains the 50s3 subunit of one 70s ribosome. the entire crystal structure4 contains two 70s ribosomes as described in remark 400.
|
| 37 | d1m0sa2 |
|
not modelled |
6.0 |
22 |
Fold:Ferredoxin-like
Superfamily:D-ribose-5-phosphate isomerase (RpiA), lid domain
Family:D-ribose-5-phosphate isomerase (RpiA), lid domain |
| 38 | d2h85a2 |
|
not modelled |
5.8 |
29 |
Fold:EndoU-like
Superfamily:EndoU-like
Family:Nsp15 C-terminal domain-like |
| 39 | d1ncsa_ |
|
not modelled |
5.7 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 40 | d1u8sa1 |
|
not modelled |
5.6 |
32 |
Fold:Ferredoxin-like
Superfamily:ACT-like
Family:Glycine cleavage system transcriptional repressor |
| 41 | d1qabe_ |
|
not modelled |
5.5 |
24 |
Fold:Lipocalins
Superfamily:Lipocalins
Family:Retinol binding protein-like |
| 42 | d1ywsa1 |
|
not modelled |
5.5 |
44 |
Fold:Rubredoxin-like
Superfamily:CSL zinc finger
Family:CSL zinc finger |
| 43 | d2ayua1 |
|
not modelled |
5.4 |
27 |
Fold:NAP-like
Superfamily:NAP-like
Family:NAP-like |
| 44 | c2ayuA_ |
|
not modelled |
5.4 |
27 |
PDB header:chaperone
Chain: A: PDB Molecule:nucleosome assembly protein;
PDBTitle: the structure of nucleosome assembly protein suggests a mechanism for2 histone binding and shuttling
|
| 45 | c1bcrA_ |
|
not modelled |
5.3 |
20 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:serine carboxypeptidase ii;
PDBTitle: complex of the wheat serine carboxypeptidase, cpdw-ii, with the2 microbial peptide aldehyde inhibitor, antipain, and arginine at room3 temperature
|
| 46 | d2yt9a1 |
|
not modelled |
5.3 |
43 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 47 | c3q3wB_ |
|
not modelled |
5.3 |
60 |
PDB header:transferase
Chain: B: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: isopropylmalate isomerase small subunit from campylobacter jejuni.
|
| 48 | c2hcuA_ |
|
not modelled |
5.3 |
60 |
PDB header:lyase
Chain: A: PDB Molecule:3-isopropylmalate dehydratase small subunit;
PDBTitle: crystal structure of smu.1381 (or leud) from streptococcus2 mutans
|
| 49 | d1x6ea2 |
|
not modelled |
5.2 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 50 | d2dlka2 |
|
not modelled |
5.2 |
57 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 51 | d2glia3 |
|
not modelled |
5.1 |
29 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
| 52 | c3kxeB_ |
|
not modelled |
5.1 |
24 |
PDB header:protein binding
Chain: B: PDB Molecule:toxin protein pare-1;
PDBTitle: a conserved mode of protein recognition and binding in a2 pard-pare toxin-antitoxin complex
|