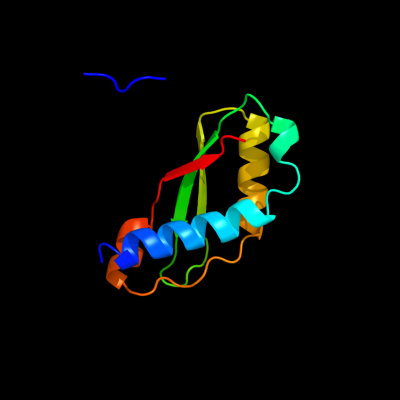
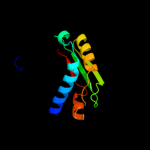
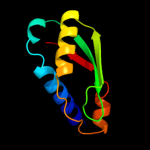
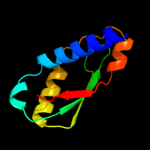
1 c1tolA_
100.0
95
PDB header: viral proteinChain: A: PDB Molecule: protein (fusion protein consisting of minor coatPDBTitle: fusion of n-terminal domain of the minor coat protein from2 gene iii in phage m13, and c-terminal domain of e. coli3 protein-tola
2 d1tola2
99.9
100
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TolA3 c2x9aB_
99.9
100
PDB header: viral proteinChain: B: PDB Molecule: membrane spanning protein, required for outer membranePDBTitle: crystal structure of g3p from phage if1 in complex with its2 coreceptor, the c-terminal domain of tola
4 d1lr0a_
99.8
16
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TolA5 d1ihra_
97.7
9
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB6 c2grxC_
97.2
15
PDB header: metal transportChain: C: PDB Molecule: protein tonb;PDBTitle: crystal structure of tonb in complex with fhua, e. coli2 outer membrane receptor for ferrichrome
7 c1xx3A_
97.2
11
PDB header: transport proteinChain: A: PDB Molecule: tonb protein;PDBTitle: solution structure of escherichia coli tonb-ctd
8 d2gskb1
96.9
9
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB9 c2k9kA_
96.7
20
PDB header: metal transportChain: A: PDB Molecule: tonb2;PDBTitle: molecular characterization of the tonb2 protein from vibrio2 anguillarum
10 d1u07a_
96.6
9
Fold: TolA/TonB C-terminal domainSuperfamily: TolA/TonB C-terminal domainFamily: TonB11 c2dzkA_
54.3
23
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: ubx domain-containing protein 2;PDBTitle: structure of the ubx domain in mouse ubx domain-containing2 protein 2
12 c1y4cA_
50.4
17
PDB header: de novo proteinChain: A: PDB Molecule: maltose binding protein fused with designedPDBTitle: designed helical protein fusion mbp
13 c2kxjA_
47.2
22
PDB header: protein bindingChain: A: PDB Molecule: ubx domain-containing protein 4;PDBTitle: solution structure of ubx domain of human ubxd2 protein
14 d2ba0g2
41.0
16
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like15 d1r4xa2
38.4
16
Fold: Subdomain of clathrin and coatomer appendage domainSuperfamily: Subdomain of clathrin and coatomer appendage domainFamily: Coatomer appendage domain16 d2e7ga1
35.4
11
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA17 c2ytxA_
35.3
36
PDB header: rna binding proteinChain: A: PDB Molecule: cold shock domain-containing protein e1;PDBTitle: solution structure of the second cold-shock domain of the human2 kiaa0885 protein (unr protein)
18 d2nn6c2
35.1
19
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like19 c2zfdB_
33.6
11
PDB header: signaling protein/transferaseChain: B: PDB Molecule: putative uncharacterized protein t20l15_90;PDBTitle: the crystal structure of plant specific calcium binding protein atcbl22 in complex with the regulatory domain of atcipk14
20 c1ybxA_
32.0
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved hypothetical protein;PDBTitle: conserved hypothetical protein cth-383 from clostridium thermocellum
21 d1wj4a_
not modelled
31.8
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain22 c2kzfA_
not modelled
31.0
11
PDB header: ribosomal proteinChain: A: PDB Molecule: ribosome-binding factor a;PDBTitle: solution nmr structure of the thermotoga maritima protein tm0855 a2 putative ribosome binding factor a
23 c1r4xA_
not modelled
29.0
14
PDB header: protein transportChain: A: PDB Molecule: coatomer gamma subunit;PDBTitle: crystal structure analys of the gamma-copi appendage domain
24 d2nn6e2
not modelled
28.3
10
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like25 c2vbcA_
not modelled
27.8
11
PDB header: hydrolaseChain: A: PDB Molecule: dengue 4 ns3 full-length protein;PDBTitle: crystal structure of the ns3 protease-helicase from dengue2 virus
26 c1pzdA_
not modelled
25.4
13
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: coatomer gamma subunit;PDBTitle: structural identification of a conserved appendage domain2 in the carboxyl-terminus of the copi gamma-subunit.
27 c3mfbA_
not modelled
21.8
33
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the s-type pyocin domain of eca16692 protein from erwinia carotovora, northeast structural3 genomics consortium target ewr82c
28 c2dnrA_
not modelled
20.0
20
PDB header: rna binding proteinChain: A: PDB Molecule: synaptojanin-1;PDBTitle: solution structure of rna binding domain in synaptojanin 1
29 c2zv4O_
not modelled
19.7
10
PDB header: structural proteinChain: O: PDB Molecule: major vault protein;PDBTitle: the structure of rat liver vault at 3.5 angstrom resolution
30 c3htyC_
not modelled
19.2
15
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein bt_0869;PDBTitle: crystal structure of hypothetical protein bt_0869 from bacteroides2 thetaiotaomicron vpi-5482 (np_809782.1) at 1.95 a resolution
31 c3mlpE_
not modelled
18.8
11
PDB header: transcription/dnaChain: E: PDB Molecule: transcription factor coe1;PDBTitle: early b-cell factor 1 (ebf1) bound to dna
32 c1dxzA_
not modelled
18.6
26
PDB header: transmembrane proteinChain: A: PDB Molecule: acetylcholine receptor protein, alpha chain;PDBTitle: m2 transmembrane segment of alpha-subunit of nicotinic2 acetylcholine receptor from torpedo californica, nmr, 203 structures
33 d1jbia_
not modelled
18.4
12
Fold: LCCL domainSuperfamily: LCCL domainFamily: LCCL domain34 c2i1jA_
not modelled
17.4
17
PDB header: cell adhesion, membrane proteinChain: A: PDB Molecule: moesin;PDBTitle: moesin from spodoptera frugiperda at 2.1 angstroms resolution
35 d2dyja1
not modelled
16.5
19
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA36 c3ol0C_
not modelled
16.3
17
PDB header: de novo proteinChain: C: PDB Molecule: de novo designed monomer trefoil-fold sub-domain whichPDBTitle: crystal structure of monofoil-4p homo-trimer: de novo designed monomer2 trefoil-fold sub-domain which forms homo-trimer assembly
37 d2nn6a2
not modelled
15.4
25
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like38 c2kncB_
not modelled
15.1
11
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
39 d2cjoa_
not modelled
15.0
24
Fold: beta-Grasp (ubiquitin-like)Superfamily: 2Fe-2S ferredoxin-likeFamily: 2Fe-2S ferredoxin-related40 d1wg1a_
not modelled
14.0
33
Fold: Ferredoxin-likeSuperfamily: RNA-binding domain, RBDFamily: Canonical RBD41 d1kkga_
not modelled
14.0
12
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA42 d1j8ba_
not modelled
13.4
10
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like43 d2je6a2
not modelled
12.8
20
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like44 d1s48a_
not modelled
11.7
22
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase45 c2rmzA_
not modelled
11.4
11
PDB header: cell adhesionChain: A: PDB Molecule: integrin beta-3;PDBTitle: bicelle-embedded integrin beta3 transmembrane segment
46 d1kyfa2
not modelled
11.3
12
Fold: Subdomain of clathrin and coatomer appendage domainSuperfamily: Subdomain of clathrin and coatomer appendage domainFamily: Clathrin adaptor appendage, alpha and beta chain-specific domain47 c3pn1A_
not modelled
11.2
20
PDB header: ligase/ligase inhibitorChain: A: PDB Molecule: dna ligase;PDBTitle: novel bacterial nad+-dependent dna ligase inhibitors with broad2 spectrum potency and antibacterial efficacy in vivo
48 d2ux9a1
not modelled
11.1
23
Fold: Dodecin subunit-likeSuperfamily: Dodecin-likeFamily: Dodecin-like49 c1sfeA_
not modelled
10.9
21
PDB header: dna-binding proteinChain: A: PDB Molecule: ada o6-methylguanine-dna methyltransferase;PDBTitle: ada o6-methylguanine-dna methyltransferase from escherichia coli
50 c1x65A_
not modelled
10.7
22
PDB header: rna binding proteinChain: A: PDB Molecule: unr protein;PDBTitle: solution structure of the third cold-shock domain of the human2 kiaa0885 protein (unr protein)
51 d2k8ea1
not modelled
10.4
14
Fold: YegP-likeSuperfamily: YegP-likeFamily: YegP-like52 d1x6oa2
not modelled
10.2
27
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like53 d1nkza_
not modelled
9.7
15
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits54 c3ecyA_
not modelled
9.6
19
PDB header: hydrolaseChain: A: PDB Molecule: cg4584-pa, isoform a (bcdna.ld08534);PDBTitle: crystal structural analysis of drosophila melanogaster dutpase
55 d1sfea1
not modelled
9.6
21
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domainFamily: Methylated DNA-protein cysteine methyltransferase, C-terminal domain56 d2cr5a1
not modelled
9.5
20
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain57 d1tl2a_
not modelled
9.5
14
Fold: 5-bladed beta-propellerSuperfamily: Tachylectin-2Family: Tachylectin-258 c3ajbB_
not modelled
9.1
42
PDB header: protein transportChain: B: PDB Molecule: peroxisomal biogenesis factor 19;PDBTitle: crystal structure of human pex3p in complex with n-terminal pex19p2 peptide
59 d1h8ca_
not modelled
9.0
27
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain60 c1cu1B_
not modelled
9.0
28
PDB header: hydrolaseChain: B: PDB Molecule: protein (protease/helicase ns3);PDBTitle: crystal structure of an enzyme complex from hepatitis c2 virus
61 d1pa4a_
not modelled
9.0
10
Fold: Alpha-lytic protease prodomain-likeSuperfamily: Ribosome-binding factor A, RbfAFamily: Ribosome-binding factor A, RbfA62 c1ky6A_
not modelled
8.9
12
PDB header: endocytosis/exocytosisChain: A: PDB Molecule: alpha-adaptin c;PDBTitle: ap-2 clathrin adaptor alpha-appendage in complex with epsin2 dpw peptide
63 d1puga_
not modelled
8.9
3
Fold: YbaB-likeSuperfamily: YbaB-likeFamily: YbaB-like64 d1v9pa3
not modelled
8.8
15
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase65 d1ccwb_
not modelled
8.7
18
Fold: TIM beta/alpha-barrelSuperfamily: Cobalamin (vitamin B12)-dependent enzymesFamily: Glutamate mutase, large subunit66 c3onrI_
not modelled
8.7
20
PDB header: metal binding proteinChain: I: PDB Molecule: protein transport protein sece2;PDBTitle: crystal structure of the calcium chelating immunodominant antigen,2 calcium dodecin (rv0379),from mycobacterium tuberculosis with a novel3 calcium-binding site
67 d1n1ba1
not modelled
8.6
23
Fold: alpha/alpha toroidSuperfamily: Terpenoid cyclases/Protein prenyltransferasesFamily: Terpenoid cyclase N-terminal domain68 c2zsjB_
not modelled
8.6
10
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
69 d1i42a_
not modelled
8.5
23
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: UBX domain70 c2k59B_
not modelled
8.5
33
PDB header: transport proteinChain: B: PDB Molecule: neuronal acetylcholine receptor subunit beta-2;PDBTitle: nmr structures of the second transmembrane domain of the2 neuronal acetylcholine receptor beta 2 subunit
71 d3bzka5
not modelled
8.5
21
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like72 d1ta8a_
not modelled
8.3
22
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase73 c3p2oB_
not modelled
8.3
11
PDB header: oxidoreductase, hydrolaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
74 c2jpxA_
not modelled
8.2
14
PDB header: viral proteinChain: A: PDB Molecule: vpu protein;PDBTitle: a18h vpu tm structure in lipid bilayers
75 c2l7qA_
not modelled
8.2
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of conjugate transposon protein bvu_1572(27-2 141) from bacteroides vulgatus, northeast structural genomics3 consortium target bvr155
76 c1zbtA_
not modelled
8.0
16
PDB header: translationChain: A: PDB Molecule: peptide chain release factor 1;PDBTitle: crystal structure of peptide chain release factor 1 (rf-1) (smu.1085)2 from streptococcus mutans at 2.34 a resolution
77 c1s1hJ_
not modelled
8.0
13
PDB header: ribosomeChain: J: PDB Molecule: 40s ribosomal protein s20;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from2 yeast obtained by docking atomic models for rna and protein3 components into a 11.7 a cryo-em map. this file, 1s1h,4 contains 40s subunit. the 60s ribosomal subunit is in file5 1s1i.
78 c2kncA_
not modelled
8.0
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
79 d1ijda_
not modelled
7.9
15
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits80 c2ihr1_
not modelled
7.9
12
PDB header: translationChain: 1: PDB Molecule: peptide chain release factor 2;PDBTitle: rf2 of thermus thermophilus
81 d1j3wa_
not modelled
7.9
22
Fold: Profilin-likeSuperfamily: Roadblock/LC7 domainFamily: Roadblock/LC7 domain82 c3p2oA_
not modelled
7.8
11
PDB header: oxidoreductase, hydrolaseChain: A: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
83 c3muxB_
not modelled
7.6
21
PDB header: lyaseChain: B: PDB Molecule: putative 4-hydroxy-2-oxoglutarate aldolase;PDBTitle: the crystal structure of a putative 4-hydroxy-2-oxoglutarate aldolase2 from bacillus anthracis to 1.45a
84 d1oysa1
not modelled
7.6
15
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: Ribonuclease PH domain 1-like85 c2l3bA_
not modelled
7.5
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein found in conjugate transposon;PDBTitle: solution nmr structure of the bt_0084 lipoprotein from bacteroides2 thetaiotaomicron, northeast structural genomics consortium target3 btr376
86 c3oqtP_
not modelled
7.4
16
PDB header: flavoproteinChain: P: PDB Molecule: rv1498a protein;PDBTitle: crystal structure of rv1498a protein from mycobacterium tuberculosis
87 c2y69R_
not modelled
7.3
33
PDB header: electron transportChain: R: PDB Molecule: cytochrome c oxidase subunit 5a;PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular2 oxygen
88 d1v54e_
not modelled
7.3
33
Fold: alpha-alpha superhelixSuperfamily: Cytochrome c oxidase subunit EFamily: Cytochrome c oxidase subunit E89 c1a4iB_
not modelled
7.1
17
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase /PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
90 d1vk8a_
not modelled
7.1
18
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: MTH1187-like91 d1dxwa_
not modelled
7.1
31
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases92 d1b04a_
not modelled
7.1
15
Fold: ATP-graspSuperfamily: DNA ligase/mRNA capping enzyme, catalytic domainFamily: Adenylation domain of NAD+-dependent DNA ligase93 c1a11A_
not modelled
7.0
30
PDB header: acetylcholine receptorChain: A: PDB Molecule: acetylcholine receptor m2;PDBTitle: nmr structure of membrane spanning segment 2 of the2 acetylcholine receptor in dpc micelles, 10 structures
94 c1yq3C_
not modelled
7.0
29
PDB header: oxidoreductaseChain: C: PDB Molecule: succinate dehydrogenase cytochrome b, large subunit;PDBTitle: avian respiratory complex ii with oxaloacetate and ubiquinone
95 c1eq8B_
not modelled
7.0
30
PDB header: signaling proteinChain: B: PDB Molecule: acetylcholine receptor protein;PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
96 c1eq8D_
not modelled
7.0
30
PDB header: signaling proteinChain: D: PDB Molecule: acetylcholine receptor protein;PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
97 c1eq8C_
not modelled
7.0
30
PDB header: signaling proteinChain: C: PDB Molecule: acetylcholine receptor protein;PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
98 c1eq8E_
not modelled
7.0
30
PDB header: signaling proteinChain: E: PDB Molecule: acetylcholine receptor protein;PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment
99 c1eq8A_
not modelled
7.0
30
PDB header: signaling proteinChain: A: PDB Molecule: acetylcholine receptor protein;PDBTitle: three-dimensional structure of the pentameric helical2 bundle of the acetylcholine receptor m2 transmembrane3 segment