1 c1vlyA_
100.0
97
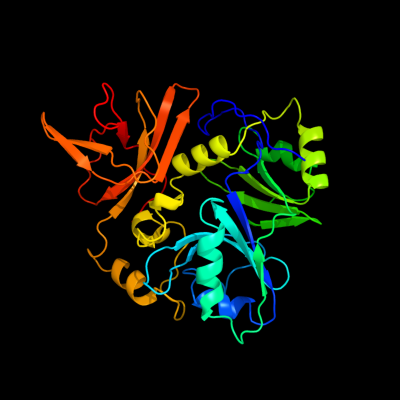
PDB header: transferaseChain: A: PDB Molecule: unknown protein from 2d-page;PDBTitle: crystal structure of a putative aminomethyltransferase (ygfz) from2 escherichia coli at 1.30 a resolution
2 c3girA_
100.0
19
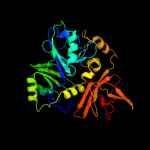
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of glycine cleavage system2 aminomethyltransferase t from bartonella henselae
3 c1vloA_
100.0
15
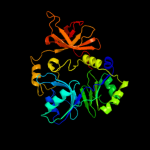
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of aminomethyltransferase (t protein;2 tetrahydrofolate-dependent) of glycine cleavage system (np417381)3 from escherichia coli k12 at 1.70 a resolution
4 c1pj6A_
100.0
15
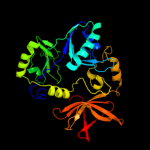
PDB header: oxidoreductaseChain: A: PDB Molecule: n,n-dimethylglycine oxidase;PDBTitle: crystal structure of dimethylglycine oxidase of arthrobacter2 globiformis in complex with folic acid
5 d1vlya2
100.0
98
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: Aminomethyltransferase folate-binding domain6 c1yx2B_
100.0
19
PDB header: transferaseChain: B: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of the probable aminomethyltransferase2 from bacillus subtilis
7 c1v5vA_
100.0
16
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of a component of glycine cleavage system: t-protein2 from pyrococcus horikoshii ot3 at 1.5 a resolution
8 c1worA_
100.0
16
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of t-protein of the glycine cleavage2 system
9 c1x31A_
100.0
16
PDB header: oxidoreductaseChain: A: PDB Molecule: sarcosine oxidase alpha subunit;PDBTitle: crystal structure of heterotetrameric sarcosine oxidase from2 corynebacterium sp. u-96
10 c1wsrA_
100.0
16
PDB header: transferaseChain: A: PDB Molecule: aminomethyltransferase;PDBTitle: crystal structure of human t-protein of glycine cleavage2 system
11 c3tfhB_
100.0
13
PDB header: transferaseChain: B: PDB Molecule: gcvt-like aminomethyltransferase protein;PDBTitle: dmsp-dependent demethylase from p. ubique - apo
12 d1vloa2
100.0
17
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: Aminomethyltransferase folate-binding domain13 d1pj5a4
100.0
15
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: Aminomethyltransferase folate-binding domain14 d1v5va2
100.0
16
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: Aminomethyltransferase folate-binding domain15 d1wosa2
100.0
15
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: Aminomethyltransferase folate-binding domain16 c2gagC_
99.8
9
PDB header: oxidoreductaseChain: C: PDB Molecule: heterotetrameric sarcosine oxidase gamma-subunit;PDBTitle: heteroteterameric sarcosine: structure of a diflavin metaloenzyme at2 1.85 a resolution
17 d1vlya1
99.5
96
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain18 d1v5va1
99.1
14
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain19 d1wosa1
99.0
19
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain20 d1vloa1
98.9
7
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain21 d1pj5a1
not modelled
98.8
12
Fold: Elongation factor/aminomethyltransferase common domainSuperfamily: Aminomethyltransferase beta-barrel domainFamily: Aminomethyltransferase beta-barrel domain22 c1xzqA_
not modelled
95.3
15
PDB header: hydrolaseChain: A: PDB Molecule: probable trna modification gtpase trme;PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
23 c3gehA_
not modelled
95.0
11
PDB header: hydrolaseChain: A: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from nostoc in complex with gdp, folinic2 acid and zn
24 c1xzqB_
not modelled
94.3
15
PDB header: hydrolaseChain: B: PDB Molecule: probable trna modification gtpase trme;PDBTitle: structure of the gtp-binding protein trme from thermotoga2 maritima complexed with 5-formyl-thf
25 d1xzpa3
not modelled
94.0
15
Fold: Folate-binding domainSuperfamily: Folate-binding domainFamily: TrmE formyl-THF-binding domain26 c3geiB_
not modelled
93.9
14
PDB header: hydrolaseChain: B: PDB Molecule: trna modification gtpase mnme;PDBTitle: crystal structure of mnme from chlorobium tepidum in complex2 with gcp
27 d3bzka5
not modelled
58.3
15
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like28 d1w4ma_
not modelled
25.3
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain29 c2ysrA_
not modelled
16.4
40
PDB header: signaling proteinChain: A: PDB Molecule: dep domain-containing protein 1;PDBTitle: solution structure of the dep domain from human dep domain-2 containing protein 1
30 d2bhua2
not modelled
14.9
15
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: alpha-Amylases, C-terminal beta-sheet domain31 c3brcA_
not modelled
13.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: conserved protein of unknown function;PDBTitle: crystal structure of a conserved protein of unknown function from2 methanobacterium thermoautotrophicum
32 d2hjsa1
not modelled
13.0
28
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain33 d1uhwa_
not modelled
12.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain34 c2vqaC_
not modelled
12.1
8
PDB header: metal-binding proteinChain: C: PDB Molecule: sll1358 protein;PDBTitle: protein-folding location can regulate mn versus cu- or zn-2 binding. crystal structure of mnca.
35 c1nnoA_
not modelled
10.9
15
PDB header: oxidoreductaseChain: A: PDB Molecule: nitrite reductase;PDBTitle: conformational changes occurring upon no binding in nitrite2 reductase from pseudomonas aeruginosa
36 d1fsha_
not modelled
10.1
20
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain37 d1xqba_
not modelled
9.8
15
Fold: YaeB-likeSuperfamily: YaeB-likeFamily: YaeB-like38 c2imuA_
not modelled
9.7
58
PDB header: viral proteinChain: A: PDB Molecule: structural polyprotein (pp) p1;PDBTitle: nmr structure of pep46 from the infectious bursal disease2 virus (ibdv) in dodecylphosphocholin (dpc).
39 d2csoa1
not modelled
9.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain40 c2gw4C_
not modelled
7.9
50
PDB header: luminescent proteinChain: C: PDB Molecule: kaede;PDBTitle: crystal structure of stony coral fluorescent protein kaede, red form
41 c2qlcC_
not modelled
7.2
21
PDB header: dna binding proteinChain: C: PDB Molecule: dna repair protein radc homolog;PDBTitle: the crystal structure of dna repair protein radc from chlorobium2 tepidum tls
42 d1uika2
not modelled
6.6
10
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Germin/Seed storage 7S protein43 d2o3la1
not modelled
6.5
23
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like44 d1v3fa_
not modelled
6.4
14
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain45 d1o7fa1
not modelled
6.1
22
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: DEP domain46 c2oceA_
not modelled
6.1
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein pa5201;PDBTitle: crystal structure of tex family protein pa5201 from2 pseudomonas aeruginosa
47 c2nv4A_
not modelled
5.9
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0066 protein af_0241;PDBTitle: crystal structure of upf0066 protein af0241 in complex with2 s-adenosylmethionine. northeast structural genomics3 consortium target gr27
48 c2nyiB_
not modelled
5.9
14
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: unknown protein;PDBTitle: crystal structure of an unknown protein from galdieria2 sulphuraria
49 d1mb4a1
not modelled
5.6
14
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain50 d1u8sa1
not modelled
5.4
21
Fold: Ferredoxin-likeSuperfamily: ACT-likeFamily: Glycine cleavage system transcriptional repressor51 d1vqod1
not modelled
5.3
19
Fold: RL5-likeSuperfamily: RL5-likeFamily: Ribosomal protein L552 d2hh6a1
not modelled
5.2
14
Fold: Left-handed superhelixSuperfamily: BH3980-likeFamily: BH3980-like53 c2a56A_
not modelled
5.1
50
PDB header: luminescent proteinChain: A: PDB Molecule: gfp-like non-fluorescent chromoprotein fp595 chain 1;PDBTitle: fluorescent protein asfp595, a143s, on-state, 5min irradiation