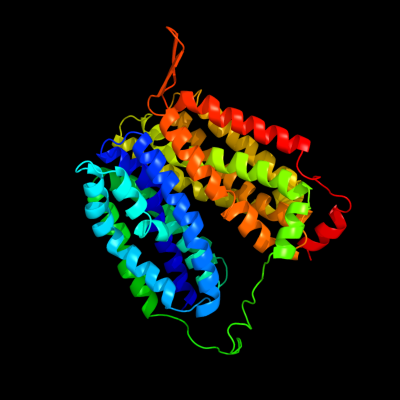
| 1 | d1pv7a_
|
|
 |
100.0 |
12 |
Fold:MFS general substrate transporter
Superfamily:MFS general substrate transporter
Family:LacY-like proton/sugar symporter |
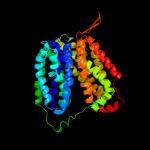
| 2 | d1pw4a_
|
|
 |
100.0 |
14 |
Fold:MFS general substrate transporter
Superfamily:MFS general substrate transporter
Family:Glycerol-3-phosphate transporter |
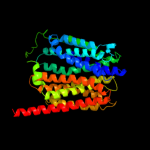
| 3 | c2gfpA_
|
|
 |
100.0 |
10 |
PDB header:membrane protein
Chain: A: PDB Molecule:multidrug resistance protein d;
PDBTitle: structure of the multidrug transporter emrd from2 escherichia coli
|
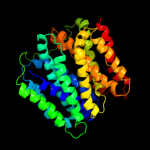
| 4 | c3o7pA_
|
|
 |
100.0 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:l-fucose-proton symporter;
PDBTitle: crystal structure of the e.coli fucose:proton symporter, fucp (n162a)
|
| 5 | c2xutC_
|
|
 |
99.9 |
13 |
PDB header:transport protein
Chain: C: PDB Molecule:proton/peptide symporter family protein;
PDBTitle: crystal structure of a proton dependent oligopeptide (pot)2 family transporter.
|
| 6 | c3qnqD_
|
|
 |
85.1 |
11 |
PDB header:membrane protein, transport protein
Chain: D: PDB Molecule:pts system, cellobiose-specific iic component;
PDBTitle: crystal structure of the transporter chbc, the iic component from the2 n,n'-diacetylchitobiose-specific phosphotransferase system
|
| 7 | c3mkuA_
|
|
 |
51.3 |
6 |
PDB header:transport protein
Chain: A: PDB Molecule:multi antimicrobial extrusion protein (na(+)/drug
PDBTitle: structure of a cation-bound multidrug and toxin compound extrusion2 (mate) transporter
|
| 8 | c3hd6A_
|
|
 |
31.1 |
9 |
PDB header:membrane protein, transport protein
Chain: A: PDB Molecule:ammonium transporter rh type c;
PDBTitle: crystal structure of the human rhesus glycoprotein rhcg
|
| 9 | c2w8aC_
|
|
 |
26.0 |
3 |
PDB header:membrane protein
Chain: C: PDB Molecule:glycine betaine transporter betp;
PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
|
| 10 | c2kncA_
|
|
 |
18.0 |
8 |
PDB header:cell adhesion
Chain: A: PDB Molecule:integrin alpha-iib;
PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
|
| 11 | d1j4na_
|
|
 |
11.9 |
14 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 12 | c1zzaA_
|
|
 |
8.9 |
12 |
PDB header:membrane protein
Chain: A: PDB Molecule:stannin;
PDBTitle: solution nmr structure of the membrane protein stannin
|
| 13 | c3a0bT_
|
|
 |
7.7 |
30 |
PDB header:electron transport
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of br-substituted photosystem ii complex
|
| 14 | c3arct_
|
|
 |
7.7 |
30 |
PDB header:electron transport, photosynthesis
Chain: T: PDB Molecule:photosystem ii reaction center protein t;
PDBTitle: crystal structure of oxygen-evolving photosystem ii at 1.9 angstrom2 resolution
|
| 15 | c2jlnA_
|
|
 |
7.5 |
11 |
PDB header:membrane protein
Chain: A: PDB Molecule:mhp1;
PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
|
| 16 | c2kluA_
|
|
 |
7.4 |
13 |
PDB header:immune system, membrane protein
Chain: A: PDB Molecule:t-cell surface glycoprotein cd4;
PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
|
| 17 | d1ymga1
|
|
 |
7.1 |
13 |
Fold:Aquaporin-like
Superfamily:Aquaporin-like
Family:Aquaporin-like |
| 18 | c1ymgA_
|
|
 |
7.1 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:lens fiber major intrinsic protein;
PDBTitle: the channel architecture of aquaporin o at 2.2 angstrom resolution
|
| 19 | c2rddB_
|
|
 |
7.0 |
8 |
PDB header:membrane protein/transport protein
Chain: B: PDB Molecule:upf0092 membrane protein yajc;
PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
|
| 20 | c3b9yA_
|
|
 |
6.9 |
11 |
PDB header:transport protein
Chain: A: PDB Molecule:ammonium transporter family rh-like protein;
PDBTitle: crystal structure of the nitrosomonas europaea rh protein
|
| 21 | c2g9pA_ |
|
not modelled |
6.4 |
25 |
PDB header:antimicrobial protein
Chain: A: PDB Molecule:antimicrobial peptide latarcin 2a;
PDBTitle: nmr structure of a novel antimicrobial peptide, latarcin 2a,2 from spider (lachesana tarabaevi) venom
|
| 22 | d2fnoa1 |
|
not modelled |
6.3 |
29 |
Fold:GST C-terminal domain-like
Superfamily:GST C-terminal domain-like
Family:Glutathione S-transferase (GST), C-terminal domain |
| 23 | c2rm9A_ |
|
not modelled |
6.0 |
19 |
PDB header:neuropeptide
Chain: A: PDB Molecule:astressin2b;
PDBTitle: astressin2b
|
| 24 | c3rl0j_ |
|
not modelled |
5.8 |
27 |
PDB header:membrane protein/exocytosis
Chain: J: PDB Molecule:syntaxin-1a;
PDBTitle: truncated snare complex with complexin (p1)
|