 |
| ||||||
| Protein Homology/analogY Recognition Engine V 2.0 | |||||||
|
 |
| ||||||
| Protein Homology/analogY Recognition Engine V 2.0 | |||||||
|
BackPhyre |
 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
What it is Instead of predicting the 3D structure of a protein sequence, often users have a solved structure and they are interested in determining if there is a related structure in a genome of interest. I have received frequent requests for this functionality and have called it "BackPhyre" to indicate how Phyre is being used in reverse.
IMPORTANT: For this procedure to work, only single protein chains must be submitted. There are currently no checks performed on your uploaded structure, so please be sure to only submit one chain. The following genomes are currently available to search If you want to search a genome not on this list please let me know. It takes considerable computation to process a large genome for use by BackPhyre so it may take a while between you asking for a genome and getting it up and running. I will maintain a list here and on the Phyre Google Group of requests and and genomes currently being processed Results and interpretation Unlike ordinary Phyre results, BackPhyre does not produce a 3D model. Instead it displays a ranked list of hits from the genomes searched, and links to the alignments. See figures below. | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
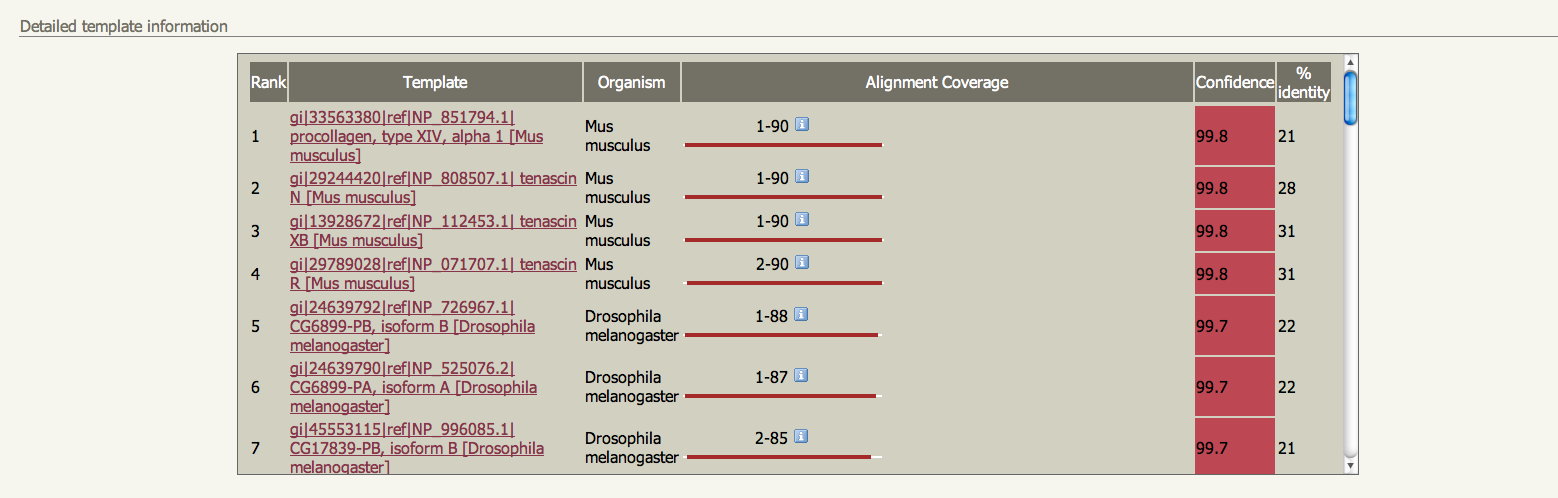
If you find an interesting hit simple retrieve the sequence using the links in the main page and submit that sequence for a normal Phyre modelling run.
Phyre is now FREE for commercial users!
All images and data generated by Phyre2 are free to use in any publication with acknowledgement
Accessibility Statement| Please cite: Phyre2.2: A Community Resource for Template-based Protein Structure Prediction | |||||||||
| Powell HR et al. Journal of Molecular Biology (2025) in press DOI: https://doi.org/10.1016/j.jmb.2025.168960 | |||||||||
|
 |
|
|||||||