|
|
 |
| Summary |
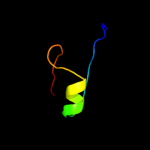
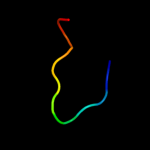
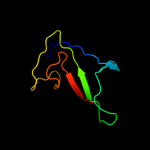
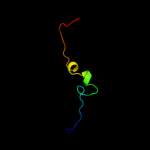
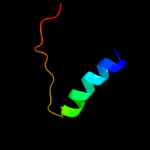
| Top model | |||||||||||||||||
| |||||||||||||||||
| Sequence analysis |
| Secondary structure and disorder prediction |
| 1 | . | . | . | . | . | . | . | . | 10 | . | . | . | . | . | . | . | . | . | 20 | . | . | . | . | . | . | . | . | . | 30 | . | . | . | . | . | . | . | . | . | 40 | . | . | . | . | . | . | . | . | . | 50 | . | . | . | . | . | . | . | . | . | 60 | |||||||||||||||||||||
| Sequence | M | I | K | T | I | L | T | I | A | L | Y | V | S | V | E | Y | C | R | G | Q | Y | F | D | Y | H | D | L | E | L | G | R | H | Y | T | N | L | T | T | K | G | D | L | V | S | D | I | Y | R | V | V | V | D | T | K | N | P | V | K | I | I | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | ||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | ? | ? | ? | ? | ? | ? | ? | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 70 | . | . | . | . | . | . | . | . | . | 80 | . | . | . | . | . | . | . | . | . | 90 | . | . | . | . | . | . | . | . | . | 100 | . | . | . | . | . | . | . | . | . | 110 | . | . | . | . | . | . | . | . | . | 120 | |||||||||||||||||||||
| Sequence | I | S | K | I | M | F | V | Y | L | Q | P | P | A | I | R | V | S | V | A | S | E | N | A | S | L | K | H | I | L | A | V | T | V | V | R | G | K | T | V | H | N | I | A | L | S | R | L | Q | S | H | R | N | K | Q | Y | E | Y | W | F | A | ||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | |||||||||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| . | . | . | . | . | . | . | . | . | 130 | . | . | . | . | . | . | . | . | . | 140 | . | . | . | . | . | . | . | . | . | 150 | . | . | . | . | . | . | . | . | . | 160 | . | . | . | . | . | . | . | . | . | 170 | . | . | . | . | . | . | . | ||||||||||||||||||||||||
| Sequence | T | D | T | L | C | D | N | D | R | S | I | E | Q | R | G | Q | P | V | H | V | S | V | S | S | I | L | P | S | K | F | G | L | L | V | K | S | V | E | N | F | N | I | G | Y | V | T | V | A | K | K | I | I | L | T | I | I | N | |||||||||||||||||||||||
| Secondary structure |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  |  | ||||||||||||||||||||||||||||||||||||||||||
| SS confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | ? | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Disorder confidence | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
| ||||||||||||||||||||||||||||||
| Domain analysis |
Hover over an aligned region to see model and summary info
Please note, only up to the top 20 hits are modelled to reduce computer load
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|