| 1 |
|
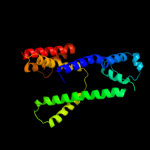
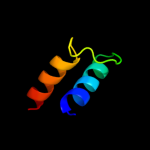
PDB 2r6g chain F domain 1
Region: 212 - 289
Aligned: 77
Modelled: 78
Confidence: 84.2%
Identity: 12%
Fold: MalF N-terminal region-like
Superfamily: MalF N-terminal region-like
Family: MalF N-terminal region-like
Phyre2
| 2 |
|
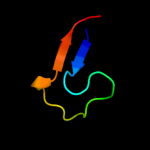
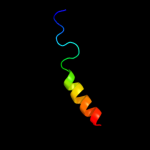
PDB 2qks chain A
Region: 226 - 313
Aligned: 88
Modelled: 88
Confidence: 41.0%
Identity: 10%
PDB header:metal transport
Chain: A: PDB Molecule:kir3.1-prokaryotic kir channel chimera;
PDBTitle: crystal structure of a kir3.1-prokaryotic kir channel chimera
Phyre2
| 3 |
|
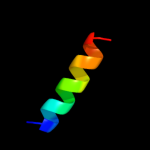
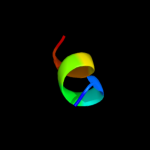
PDB 1m8l chain A
Region: 17 - 79
Aligned: 63
Modelled: 63
Confidence: 32.7%
Identity: 13%
PDB header:viral protein
Chain: A: PDB Molecule:vpr protein;
PDBTitle: nmr structure of the hiv-1 regulatory protein vpr
Phyre2
| 4 |
|
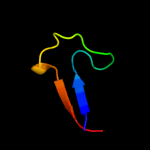
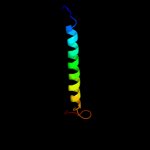
PDB 1xl6 chain B
Region: 226 - 313
Aligned: 88
Modelled: 88
Confidence: 29.7%
Identity: 6%
PDB header:metal transport
Chain: B: PDB Molecule:inward rectifier potassium channel;
PDBTitle: intermediate gating structure 2 of the inwardly rectifying k+ channel2 kirbac3.1
Phyre2
| 5 |
|
PDB 3dh4 chain A
Region: 63 - 278
Aligned: 211
Modelled: 216
Confidence: 21.5%
Identity: 15%
PDB header:transport protein
Chain: A: PDB Molecule:sodium/glucose cotransporter;
PDBTitle: crystal structure of sodium/sugar symporter with bound galactose from2 vibrio parahaemolyticus
Phyre2
| 6 |
|
PDB 3hie chain A
Region: 9 - 35
Aligned: 27
Modelled: 27
Confidence: 14.5%
Identity: 26%
PDB header:exocytosis
Chain: A: PDB Molecule:exocyst complex component sec3;
PDBTitle: structure of the membrane-binding domain of the sec3 subunit2 of the exocyst complex
Phyre2
| 7 |
|
PDB 1w8x chain P
Region: 292 - 308
Aligned: 17
Modelled: 17
Confidence: 11.1%
Identity: 12%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 8 |
|
PDB 3a58 chain A
Region: 9 - 35
Aligned: 27
Modelled: 27
Confidence: 9.9%
Identity: 26%
PDB header:protein transport/exocytosis
Chain: A: PDB Molecule:exocyst complex component sec3;
PDBTitle: crystal structure of sec3p - rho1p complex from2 saccharomyces cerevisiae
Phyre2
| 9 |
|
PDB 1bh9 chain B
Region: 28 - 65
Aligned: 36
Modelled: 38
Confidence: 8.0%
Identity: 28%
Fold: Histone-fold
Superfamily: Histone-fold
Family: TBP-associated factors, TAFs
Phyre2
| 10 |
|
PDB 3gqn chain A
Region: 7 - 30
Aligned: 24
Modelled: 24
Confidence: 6.2%
Identity: 21%
PDB header:viral protein
Chain: A: PDB Molecule:preneck appendage protein;
PDBTitle: crystal structure of the pre-mature bacteriophage phi29 gene product2 12
Phyre2
| 11 |
|
PDB 1bm4 chain A
Region: 36 - 61
Aligned: 26
Modelled: 26
Confidence: 5.9%
Identity: 19%
PDB header:viral protein
Chain: A: PDB Molecule:protein (moloney murine leukemia virus capsid);
PDBTitle: momlv capsid protein major homology region peptide analog
Phyre2
| 12 |
|
PDB 2xl1 chain A
Region: 52 - 68
Aligned: 17
Modelled: 9
Confidence: 5.2%
Identity: 24%
PDB header:translation
Chain: A: PDB Molecule:arginine attenuator peptide;
PDBTitle: structural basis of translational stalling by human cytomegalovirus2 (hcmv) and fungal arginine attenuator peptide (aap)
Phyre2
| 13 |
|
PDB 3ixz chain B
Region: 220 - 261
Aligned: 42
Modelled: 42
Confidence: 5.1%
Identity: 7%
PDB header:hydrolase
Chain: B: PDB Molecule:potassium-transporting atpase subunit beta;
PDBTitle: pig gastric h+/k+-atpase complexed with aluminium fluoride
Phyre2